Plant Hormones-Cytokinins.
Cytikinins were discovered by Skoog and Miller 1957 ; while working on the callus under in vitro conditions. They found the callus that develops from the stem explants, containing both pith and vascular elements, develops well. But the explants containing just pith cells produces callus, but further growth of its stops, even in the presence of optimal concentration of auxins. This is because the cell in the callus somehow rendered incapable of cell division. If such callus is supplanted with vascular tissues, extract of vascular tissues, coconut milk or malt extracts, the growth of the callus will be restored and the cells exhibit mitotic activity. This effect has been attributed to the presence of some active principle in the supplanted coconut milk. Cytokinins in coconut milk have various functions in association with other hormones or its accessories.
Kinetins and their effects; labs.bio.unc.edu
Auxins and Cytokinin’s- site of synthesis; www.svt.ac-diion.fr
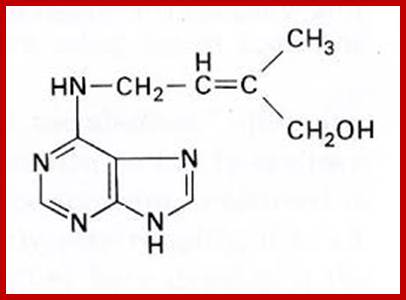
Zeatin;www.prezi.com

Gottieb Haberlandt

Skoog 1954-Kinetin; www.fagro.edu.uy

Kenneth Thimann
The above three persons are great scholars in the field of Plant hormones.
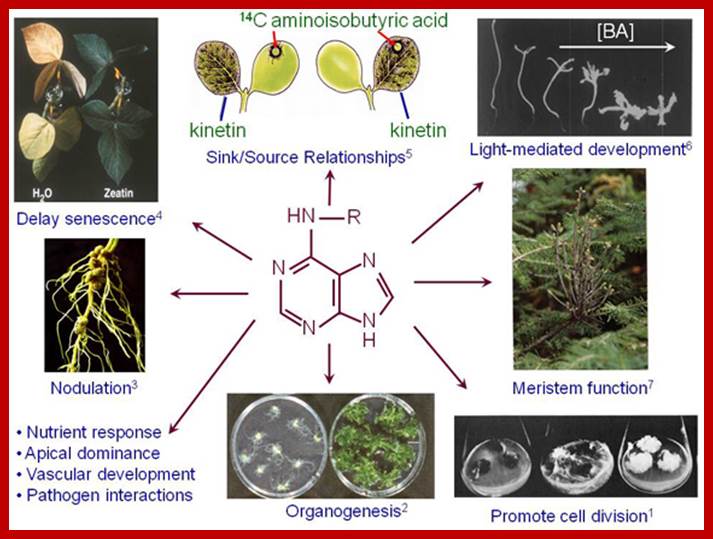
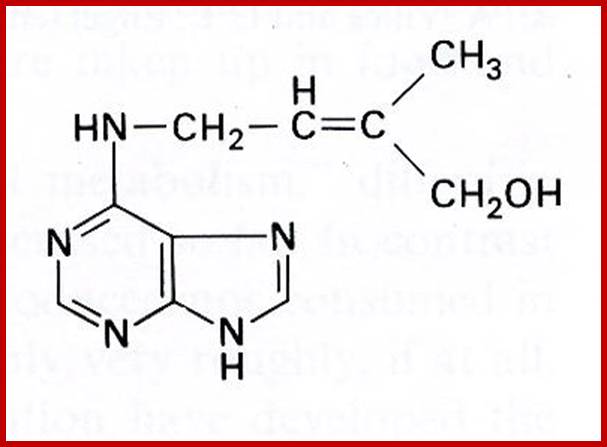
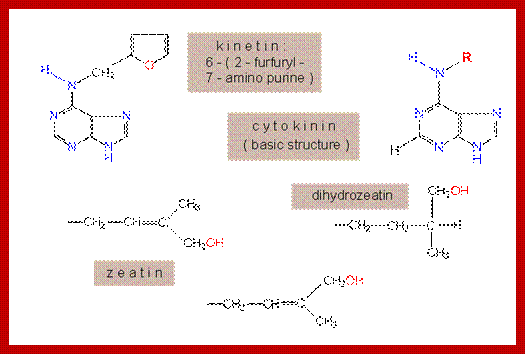
Some of the naturally occurring Cytokinin’s are 6-Furfuryl-aminopurine, Ribosyl Zeatin (immature corns), Zeatin, Isopentinyladenine and Dihydrozeatin. Interestingly, the above said compounds are also found in denatured products of nucleic acids. Many synthetic cytokinins are also available in the market, ex., 6-Benzyl amino purine, 6-Phenyl amino purine and many others. Cytokinins act on the cells that produce it- ‘Autocrine’ system. Cytokinins transiently inactivated by glycosylation at N3,N7 and N9. Cytokinins can be irreversibly degraded by oxidative cleavage. The Diphenyl urea is one of the synthetic cytokinin; Thidiazuron is a new class of cytokinin, it inhibits cytokinin oxidase that is why it is extensively used in plant tissue culture. There are 200 or more synthetic cytokinins.
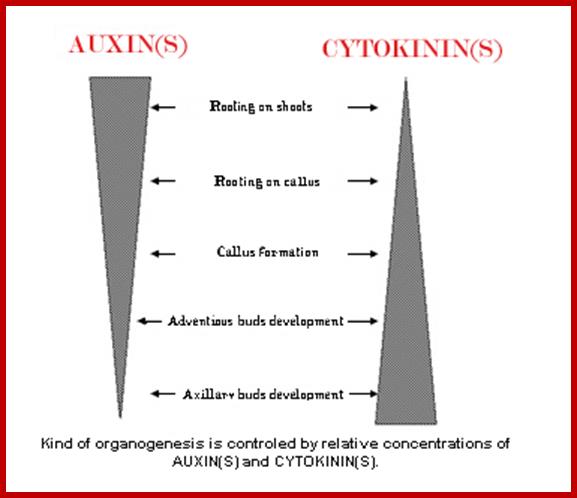
Gradient concentration Auxin and Cytokinin’s, from stem to root and from root to stem respectively;http://www2.ulg.ac.be/
www.s10.lite.msu.edu

Cytokinins; www.galleryhip.com
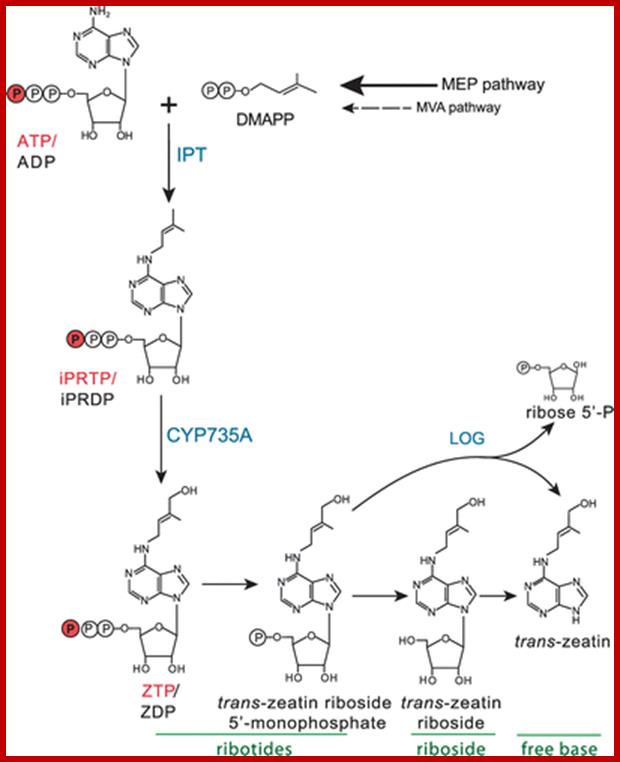
The proposed biosynthesis of trans-zeatin tri-/diphosphate in Arabidopsis is shown. Both ADP and ATP are likely substrates for the plant IPT enzyme, and these and their di- and tri-phosphate derivatives are indicted together (e. g. ATP/ADP). See text for more details. http://www.bioone.org/doi
Distribution:
Cytokinins have been detected in a wide variety of plants; from unicellular yeasts, algae and multi cellular higher plants. Particularly in higher plants, cytokinins are found in root tips, xylem, young leaves; endosperms of developing fruits, germinating seeds and tumour tissues.
Site of Synthesis:
Most of the cytokinins required for the plant body are synthesized in the root tip meristems, and then they are translocated to different regions particularly to meristematic region and expanding tissues; transportation is through xylem stream. This observation has been supported by many studies. Concentration cytokinins is ~0.1 to 10ppm/gram fresh weight. Cytokinin synthesis is not just believed to be root tips and Using MPSS (massively parallel signature sequencing) we identified 823 and 917 genes that were up- and down regulated, respectively, following 24 h of IPT induction
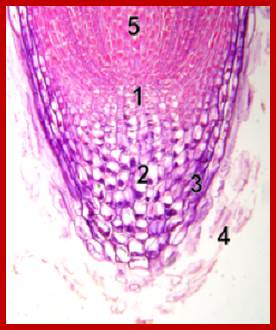
Site of CK synthesis: www.planthormones.info

www.study.com; www.education-portal.com
The active principle responsible for inducing cell division was isolated first from the extracts of yeasts. Such a substance was called kinetin, later the name was changed and called as cytokinin; kinetin terminology was misleading for another class of compounds called kinins which were already known to be found is animal systems. However, the term cytokinin has been given to all those compounds that are capable of inducing cell division in the presence of optimal concentration of auxin in plants. Now it is known that a good source for cytokinin is coconut liquid endosperm and milky endosperm of sweet corns.
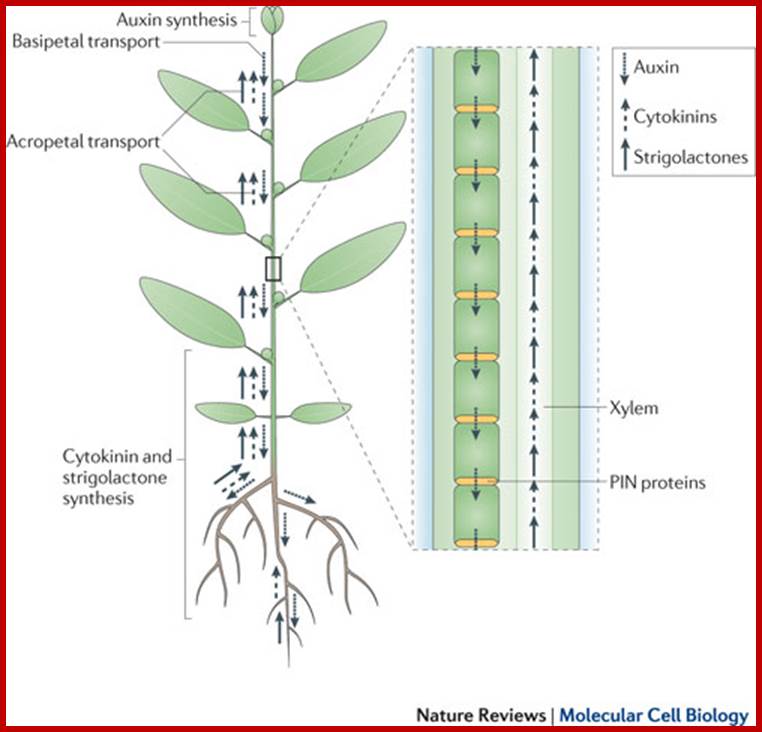
The long-distance hormonal network in shoot branching control. Auxins, cytokinins and strigolactones (or strigolactone derivatives) are three classes of hormones that are involved in the control of bud activation. These hormones are transported throughout the plant, forming a systemic network that allows integration of information between different plant organs. Auxin is mostly produced in the young expanding leaves of growing shoot apices and is actively transported basipetally in the polar auxin transport stream, involving basally localized PIN-FORMED (PIN)-type auxin efflux carriers, in particular PIN1. Strigolactones and cytokinins are mainly produced in the root, but also locally in the shoot, and are transported acropetally in the xylem. Malgorzata A. Domagalska & Ottoline Leyser;www.nature.com500
For example, the amount of cytokinin found in the excised petiole is less than the petiole that has rooted. If the root tips are cut, the growth of the stem apex is more or less inhibited till adequate supply of cytokinin is restored by new root formation. Significant amount of cytokinin is synthesized in required endosperm of palm fruits, kernel of cereal grains and others. Required Cytokinin is synthesized in root tips then it is translocated upwards in the phloem sieve tubes and distributed to different Ariel parts of the plant body.
Cytokinin biosynthesis is ATP/ADP driven isopentylatransferase IIPT genes. Seven IPT genes are found in Arabidopsis; AtLp1 and Atlp, 3, AtlP4, ArLP5 AtlP6, AtLP7 and AtLP8. The seven genes produce Cytokinin precursor-Isopentinyladenine. Two cytochrome P450 monooxygenases (CYP735A1 and CYP735A2) then catalyse the hydroxylation of Isopentinyladenine-type cytokinins. In addition, the LONELY GUY (LOG) gene family encodes enzymes that convert cytokinin from an inactive to an active form. Cytokinin degradation is mediated by the cytokinin oxidases, which were first described over 30 years ago, these enzymes, are encoded by the CYTOKININ OXIDASE (CKX) gene family.
But cytokinin uptake is not well known; however it is known purine Permease (PUP) protein has very high affinity and it binds and transport through the PUP transporters.
Almost all naturally occurring cytokinins are the products of purine nucleotide derivatives. The presence of iospentenyl adenine in some tRNAs has misled people to believe that the source of cytokinins is tRNAs but it is not the case. Nevertheless the site of synthesis of cytokinins in young and developing plants is restricted to meristematic regions of the root tips; where certain enzymes utilize purine and convert them to cytokinins. The presence of isopentenyl adenine in many tRNA is not due to the incorporation of cytokinins into tRNAs. However, the exact mechanism and biosynthetic pathway of cytokinins is yet to be elucidated.
Interaction:
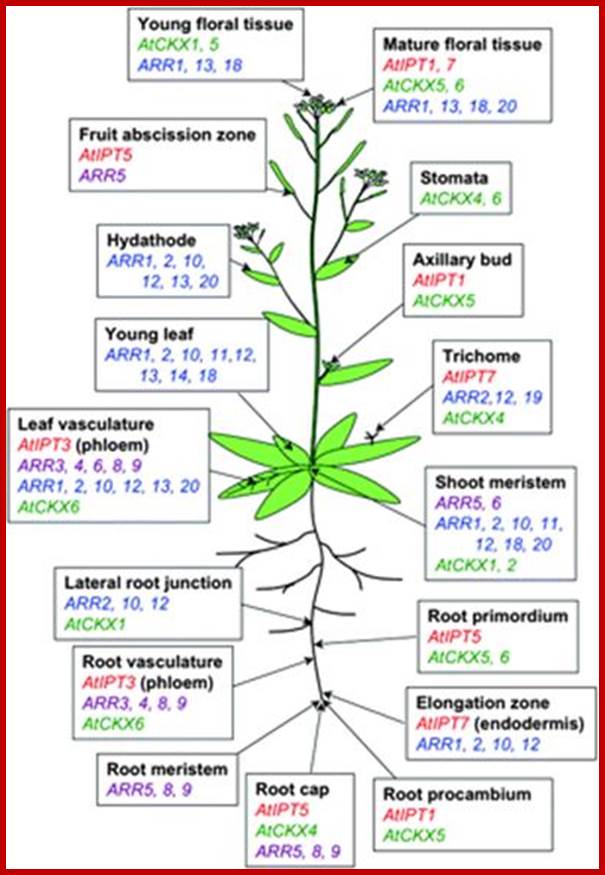
Spatial distribution of cytokinin-related gene expression in Arabidopsis. Data are based on studies of promoter:GUS (Miyawakiet al., 2004) and promoter:GFP (Takei et al., 2004a) fusions of AtIPTs (red), Type A ARRs (violet; D'Agostino et al., 2000; Kiba et al., 2002, 2003; Ferreira and Kieber, 2005; Yokoyama et al., 2007), Type B ARRs (blue; Mason et al., 2004; Tajima et al., 2004), and AtCKXs (green; Werner et al., 2003, 2006).;jxb.oxfordjournals.org
General Metabolism:
Cytokinins’ effect on respiration is very interesting. It enhances the rate of respiration in the callus, but the same hormone when applied to senescing leaves, brings down the rate of respiration but retards the degradation of chlorophyll and enhances the rate of chlorophyll synthesis. In addition it also increases the rate of metabolic activities involved in C3 pathway. In certain systems cytokinin induces nitrate reductase activity, but in callus it does not. The root nodule development in legumes and its metabolic activity is subtly regulated by the interaction between auxins and cytokinins.
Nucleic acid Synthesis:
Although cytokinin is known to stimulate cell division; it alone does not induce DNA replication as a prelude to cell division. But in the presence of auxin, it promotes DNA replication. So it is suggested that cytokinin stimulates and auxin promotes DNA replication. For example, an aged and non proliferating callus cells can be stimulated to undergo mitotic divisions by the application of cytokinin.
Cytokinin binding protein i.e. Receptor proteins have been identified in many plant systems. The cytokinin-protein complex is known to induce transcription activity in a pea bud chromatin in a cell free system. In Soybean and French been hypocotyls, it transitorily inhibits IBA induced RNA synthesis for a period of 6-7 hours, but later it stimulates RNA synthesis; this may have bearing on the inhibitory effect of Cytokinins on IBA induced new root formation.
Cytokinin induces rRNA transcription: However, the signal transduction pathways responsible for pol I regulation are poorly understood. We tested the effects of exogenously applied plant hormones on promoter-dependent rRNA transcription inArabidopsis thaliana Gibberellic acid, abscisic acid, auxin, and ethylene had no detectable effect on rRNA transcription, but kinetin (a cytokinin) stimulated rRNA transcription within 1 h of treatment. Increased steady-state levels of accurately initiated rRNA transcripts, detected by S1 nuclease protection, were paralleled by increased levels of nascent rRNA transcripts in isolated nuclei. Therefore, the primary effect of cytokinin appears to be at the level of transcription initiation rather than rRNA stability. Pol I accounts for ∼34% of total nuclear transcription in untreated plants and ∼60% following cytokinin treatment. http://www.jbc.org/
Specific species of tRNAs, containing cytokinin activity, is seen in the cotyledons and hypocotyl segments of bean plants. But this has been identified as due to post transcriptional modification of tRNA. Recent investigations have clearly elucidated that the presence of cytokinin moiety in some tRNA structures is not responsible for eliciting any cytokinin mediated responses, however it is to be noted that many tRNAs contain isopentenyl adenine (IPA) in the anticodon loop region.
Microarray meta-analysis using thirteen microarray experiments combined with empirically-defined filtering criteria identified a set of 226 genes differentially regulated by cytokinin, a subset of which have previously been validated by other methods. RNA-seq validated about 73% of the up-regulated genes identified by this meta-analysis.
RNA-seq analysis identified 73 cytokinin-regulated genes that were not represented on the ATH1 microarray; majority genes expressed used in secondary metabolism; RNA-seq analysis identified 73 cytokinin-regulated genes that were not represented on the ATH1 microarray. But the expression of CIG2 is specific to cytokinins.
BA-induced stimulation of transcription in chloroplasts- of rrn16, rrn23, rps4, rps16, rbcL, atpB, and ndhC required light during the period of preincubation and was further enhanced by light during the incubation on BA, whereas activation of transcription of trnEY, rps14, rpl16, matK,petD, and petLG depended on light during both periods. Our data reveal positive and differential effects of cytokinin on the transcription of chloroplast genes that were dependent on light and on the age (developmental stage) of cells and leaves. In cultured green cells. The expression of cig2 is specific to cytokinin and is not induced by other phytohormones. The amino acid sequence encoded by cig2 is similar to the GDP/GTP exchange factor eIF2B, which regulates translation initiation. The expression of these cig suggests a complex induction system involving cytokinin and other phytohormones.
Together with auxin, another plant hormone, cytokinins can reprogram terminally differen-tiated leaf cells into stem cells and support shoot regeneration indefinitely in plant tissue culture; Thus, cytokinins are master regulators of plant growth and development, which are highly plastic and adaptive, as well as remarkably resilient and perpetual. Using MPSS (massively parallel signature sequencing) we identified 823 and 917 genes that were up- and down-regulated, respectively, following 24 h of IPT induction. http://www.plantphysiol.org/; http://www.ncbi.nlm.nih.gov/.
Protein synthesis:
Quite a number of experiments, involvement in vivo systems or cell free invitro systems, have demonstrated that cytokinins first stimulate translational activity without any concomitant increase in the synthesis of mRNAs. This particular effect has been attributed to cytokinin’s ability to activate pre existing mRNPs and ribosomal proteins needed for chain initiation. Fox et al, have demonstrated that cytokinins bind to ribosomal surface through a receptor protein. This protein activates ribosomes by dephosphorylating specific ribosomal protein.
Furthermore, the increased activity of protein synthesis in response to cytokinin has been found to be concentration dependent. It is also interesting to note that cytokinin mediated protein synthesis; either as short term or long term effects, shows the synthesis of new proteins in treated tissues. At least in the hypocotyls of French bean, cytokinin inhibits some proteins that are induced by IBA, but cytokinin by itself enhances the rate of tubulin synthesis similar to that of IBA.
Cell Division:
The name cytokinin is derived from its ability to induce cytokinesis during cell divisions. Though cytokinins stimulate the process, the permissive effect of it is controlled by auxins. When cytokinin is provided to a liquid culture medium containing plant cells, the protein synthetic activity of the cells is greatly stimulated. Moreover, some of the proteins thus synthesized are new ones. These observations suggest that cytokinins control cytokinesis by regulating the synthesis of some specific protein factors that are required for cytokinesis. Cytokinins stimulate the activation of CDK at G2 phase of cell division. Cell cycle control is essential for the initiation and maintenance of meristems and for the regulation of organogenesis. Auxin and cytokinin hormones are implicated in cell cycle control since they strongly influence the division of cells that are in culture at G1/S and importantly G2/M phase.
Cell Enlargement:
Isolated cotyledons of Cucumis, radish and other plants expand dramatically when they are treated with cytokinin. The expansion of cotyledons is rather more due to cell enlargement than due to cell divisions. During cytokinin induced cell enlargement, respiratory activity increases significantly and greater amounts of K+ ions are accumulated in the cells. At the same time, cells in response to the hormones induce the synthesis of few minor species of RNAs and some proteins. But the inhibitors of respiration, transcription and translation completely inhibit cytokinin mediated cell enlargement. Interestingly such cotyledons also respond to red light treatment and enlarge. However the red light induced enlargement cannot be reversed by far red light treatment which further suggests that cytokinins bring about permeability changes within the membranes.
Richmond & Lang’s effect:
Senescense of leaves leads to yellowing and finally leads to the fall from the plant. If a young excised leaf is kept in water, it slowly changes its color to yellow and dies. If such leaves are provided with cytokinin, the yellowing is significantly delayed and such an effect is called Richmond and Lang’s effect; named after the discoverers.
Senescense is a common feature exhibited by parts of the plant which show definite growth pattern. With age, the structures like leaves, flowers, etc., senesce and die. Among various factors, decrease n the content of auxin acts as a very important factor in inducing senescence. In matured leaves, still attached to the plant body with time, senescence sets in and leads to the degradation of chlorophyll. Catabolic activity increases and the formation of abscission layer begin. In detached leaves also one finds similar processes. But cytokinins prevent and prolong the initiation of senescence for a quite a period of time. This effect of cytokinin has been explained as due to the prevention of degradative catabolic processes by the way of repression activity of few hydrolysing enzymes like protease, RNAse, DNAse etc. Furthermore, cytokinin facilitates the chlorophyll synthesis. It also sustains the activity of carbon fixation, RNA synthesis and protein synthesis. Still the exact mechanism by which cytokinins prevent aging and senescence is not known.
Effect on Dormancy:
Dormant buds that develop due to certain adverse environmental factors remain inactive for a long time. If such buds are treated with cytokinins they come out of dormant state and sprout. This is due to the effect of cytokinin in activating cell division which was prevented by mitotic blocks present in the dormant besides. Interestingly, cytokinins also overcome auxin imposed apical dominance and stimulate the growth of the axillary buds, probably by overcoming the factors such as mitotic blocks.
Interaction of cytokinins with auxins in morphogenesis:
Plant development is a series of biochemical events which ultimately leads to morphogenic changes. This process is regulated by a number of phytohormones which bring about differential gene activity leading to the development or specific phenotype. The interaction between various hormones is very complex and they operate at different levels like differentials transcription, protein synthesis, enzyme activity, permeability etc. Here the interaction between cytokinin and auxin id discussed briefly.
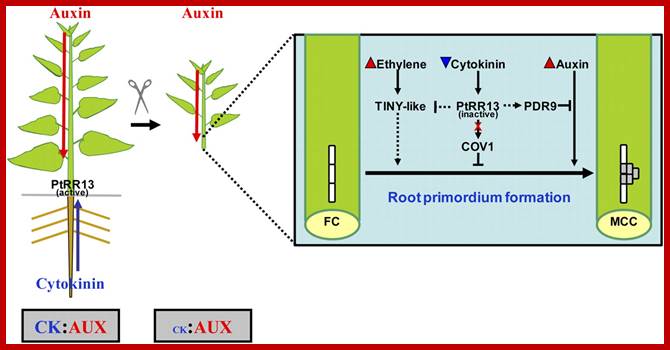

http://pixgood.com/cytokinins-in-plants.html
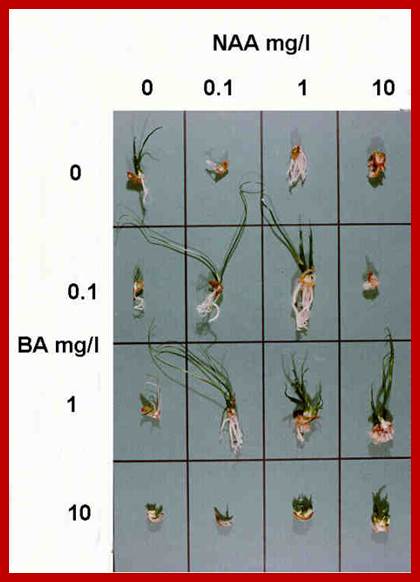
The culturing of plant explants with various combinations of cytokinins and Auxins show the inhibitory effect of cytokinin on auxin induced new root formation.
In tissue culture experiments, the plant explants are supplemented with optimal nutrients including carbohydrate as the energy source and hormones as growth regulators. Plant explants, which may be leaf segments stem segments, roots or embryos develop and produce an undifferentiated tissue called callus is a defined culture medium. This behavior is due to the presence of balanced concentration of auxin and cytokinin in the nutrient medium. The callus is like a cancerous tumour, where the cells are undifferentiated and they are under the spell of uncontrolled mitotic activity. If the concentration of auxin with respect to that of cytokinin ratio is changed, from the ratio that is favorable for the growth of only callus, the cells of the callus undergo transformation to produce organs like roots or shoots. If the ratio between auxin and cytokinin is high, the callus cells undergo transformation and produce roots. On the other hand, if the ratio between auxin and cytokinin is low the callus cells initiate shoots. As all cells in the callus are totipotent, depending upon the hormonal concentration, cellular genetic material is differentially expressed and their products induce specific morphogenetic events leading to organ formation. In this system it is the concentration of each of the hormones provide the signal for differentiation of undifferentiated cells into new organs.
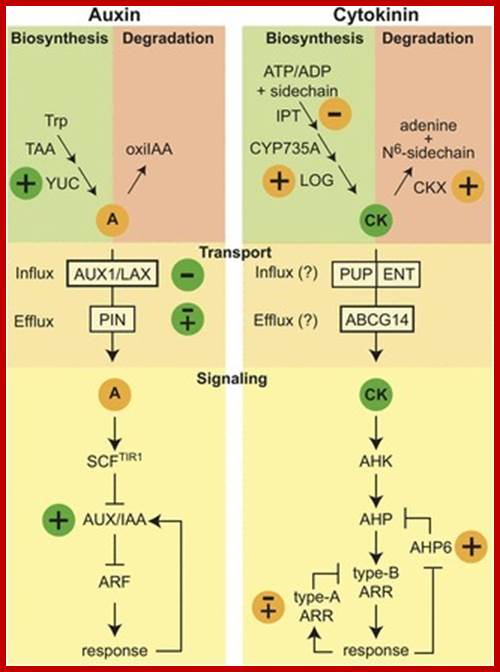
The Ying-Yang of hormonesw: cytokinins and Auxins; www.scoop.it
Inspite of having sufficient knowledge about the hormonal effects in organogenesis, the molecular basis of morphogenesis is not very clear; however, one example can be used to illustrate how auxins and cytokinins interact and modulate molecular events during morphogenesis. Adventitious root formation in the hypocotyls of phaseolus vulgaris in response to auxin treatment is a process of redifferentiation and reorganization of pericyclic cells into root primordia. While IBA induces new root formation, cytokinins inhibit IBA mediated root initiation. At molecular level both auxin and cytokinins increase the rate of protein synthesis without increase in transcriptional activity at 30 minutes of treatment. In the case of auxin treated hypocotyls increase in transcription is detected after 45-60 minutes, but not in cytokinins treated. In the hypocotyls treated with both auxin and cytokinins one does not observe such changes at short time but observed at longer periods. This observation comes from quantitating in vivo and in vitro protein synthesis using 14C leucine. Also purified poly-A is used for quantitation using in vitro translation. The results show increase in translation at later stages is due to transcriptional activation. General Protein analysis on SDS-PAGE at 24hrs, 48hr and 72 hrs, shows a remarkable increase in two bands at 55Kda and 58Kda and few other bands of high molecular weight. However increase in the said proteins was not detected in cytokinin treated segments. The bands were identified as Tubulin alpha and beta proteins. More over in Auxin treated segment discerned from in vitro translation of Pol(A) RNA shows one more 115Kds band showed up only in auxin treated but not in any others. This increase is only transitory from 24hrs to 36hrs. Then the protein band disappears. The role of this protein is not known.
At molecular level Auxin acts through Auxin response factor for activation of genes. But the activation of transcription at 36 hrs or more, by cytokinins is yet to be found. However cytokinins action of activation of genes has been discerned to some extent in Arabidopsis. Cytokinin acts as signal molecule and it binds to dimeric receptors anchored in plasma membrane. The receptors are believed to be similar to receptor tyrosine kinases (RTKs), but their kinase activity is restricted to Histidine residues (HPK), so they are called Arabidopsis histidine protein kinases (HKs). This kinase activates histidine phospho transfer proteins (AHPs) transducers. These are response regulators which can activate or repress gene expression. AHK-ps enter the nucleus and interact with ARRs (nuclear response regulators) and activate transcription. Such components were also observed in maize.
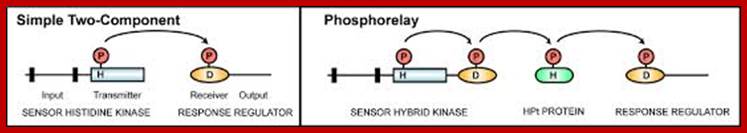
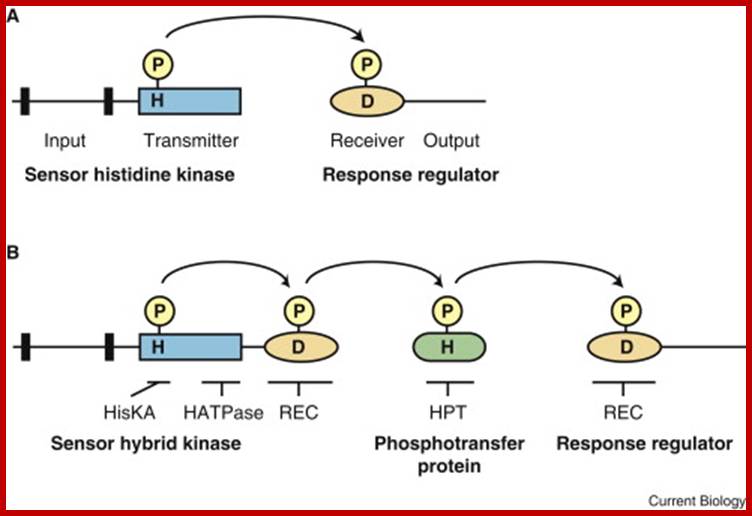
Top fig.;labs.bio.unc.edu;
Bottom fig. Elements of two-component signaling systems. www.cell.com.
Histidine kinase domains are indicated by rectangles, receiver
domains by ovals, histidine-containing phosphotransfer domains by rounded
rectangles, and transmembrane domains by black bars. Sites of phosphorylation
upon histidine (H) and aspartic acid (D) residues are indicated. (A) Simple
two-component system that employs a histidine kinase and a response
regulator. (B) Multi-step phosphorelay that employs a hybrid histidine kinase
with both histidine kinase and receiver domains, a histidine-containing
phosphotransfer protein, and a response regulator. Regions covered by
consensus domains (HisKA, HATPase_c, HPT, and REC) are indicated. http://www.cell.com/
(A) A basic prokaryotic two-component system with a sensor
histidine kinase and a response regulator. H and D represent the conserved
phosphoaccepting histidine and aspartate residues involved in phosphorelay
signaling. (B) A multistep phosphorelay system involving a hybrid
sensor kinase, with input, transmitter and receiver domains, a
histidine-containing phosphotransfer protein and a response regulator. http://www.bioone.org/
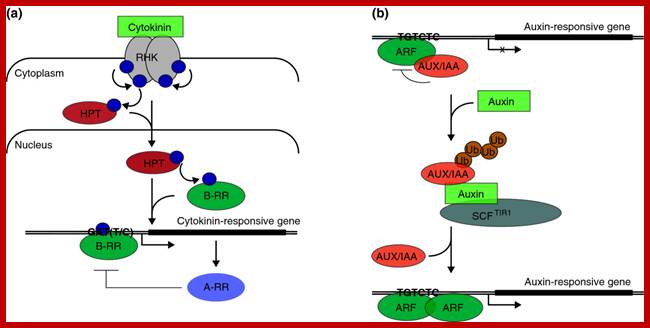
Cytokinin and auxin signaling in Arabidopsis. Single lines indicate cell membranes; double lines represent the chromosome; bent arrows indicate positions of transcription initiation. (a) Cytokinin binding to a receptor histidine protein kinase (RHK) such as AHK3 triggers kinase autophosphorylation and initiates a phosphorelay cascade [5]. The phosphoryl group (blue sphere) transfers to a receiver domain in the receptor and subsequently to a histidine phosphotransfer protein (HPT), triggering HPT translocation to the nucleus. There, the phosphorylation is relayed to an Arabidopsisresponse regulator (ARR) such as ARR1. B-type RRs (B-RR) activate transcription of cytokinin-responsive genes, some of which contain a GAT(T/C) DNA sequence motif [7]. Cytokinin-responsive A-type RRs (A-RR) act to repress cytokinin signaling. (b) Auxin signaling is based on auxin-dependent, proteasome-mediated degradation of AUX/IAA repressors (see [9] and references therein). AUX/IAA proteins dimerize with and repress the activity of transcription factors in the AUXIN RESPONSE FACTOR (ARF) family, which bind TGTCTC-containing DNA sequence elements in promoters of auxin-responsive genes. Auxin-dependent gene expression is mediated by the release of ARF proteins from AUX/IAA repression as a result of proteasome-mediated degradation of AUX/IAA proteins. Auxin serves as the switch by binding to an F-box protein such as TRANSPORT INHIBITOR RESPONSE1 (TIR1) and enhancing its interaction with AUX/IAA proteins, increasing the rate of AUX/IAA ubiquitination (Ub) by the Skp1-Cul-F-box E3 ubiquitin ligase complex SCFTIR1. http://www.genomebiology.com/
Outline of cytokinins pathways:
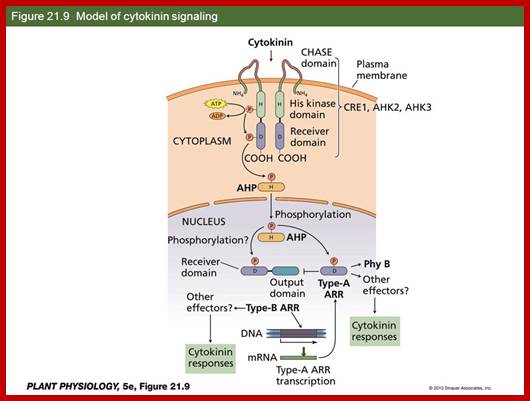
The plant hormone cytokinin is perceived by membrane-located sensor histidine kinases. Arabidopsis (Arabidopsis thaliana) possesses three cytokinin receptors: ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and CYTOKININ RESPONSE1/AHK4. Receptors are found in PM.
The large majority of cytokinin receptors are localized to the ER, suggesting a central role of this compartment in cytokinin signaling. A modified model for cytokinin signaling is proposed.
There are four steps in cytokinins signaling pathway as shown in the above diagram. AHK senses signals, AHP nuclear translocator and localizes in the nucleus and activates ARR genes and a negative feedback loop through Cytokinin inducible ARR gene products.
Arabidopsis encode three cytokinins receptors; cytokine response gene1 (CRE1 also called AHK4) ,Woodenleg (Wol) and AHK2/3. There are other histidine kinases such as CK11 and CK12 (AHK5) which are independent of cytokinins but they do respond to cytokinins. Mutants of CRE1 and Wol show defects in cytokinins’ mediated shoot formation and defects in root vasculature.
The ARR (A response regulator proteins) ( such asARR1, ARR2,and ARR10, which are transcriptional activators carry MYB like DNA binding domains, and also contain glutamine Q’ rich activating domains. ARR p-lation activates transcription of A-type ARRs. Expression of cyclin D and ARR5 show they are the major sites of cytokinins action in root and shoot meristems. There are 54 gene encode AHKs, AHPs and ARRs.

Three two-component histidine kinase proteins in Arabidopsis. CKI1 and CRE1 are similar in overall organization but quite diverged in sequence. The ETR1 protein is a representative of the ethylene receptor family. These proteins do not have an extracellular ligand binding domain since ethylene binding occurs within the hydrophobic regions [3]. The grey bars represent hydrophobic regions and the blue section is presumed to be extracellular. The GAF domain is a conserved motif that may function in cyclic-GMP binding. H and R represent the histidine kinase and response regulator domains respectively. http://www.sciencedirect.com/

Cytokinins - Cytokinins promote shoot development, delay leaf senescence, contribute to stress and pathogen responses, and serve as important signals to integrate growth rates throughout the plant. The identification of genes controlling CK synthesis and signaling has allowed us to understand the molecular basis for some of these effects with the hope of manipulating them for improved crop productivity, through increased drought tolerance, nutrient uptake and seed production. Furthermore, the exploration of the histidine-kinase phosphorelay system that is central to CK signaling has revealed a complex and fascinating cell signaling pathway; John Chandler (john.chandler@uni-koeln.de) Cytokinin’s signal transduction leading to gene activation; http://molbio.mgh.harvard.edu/
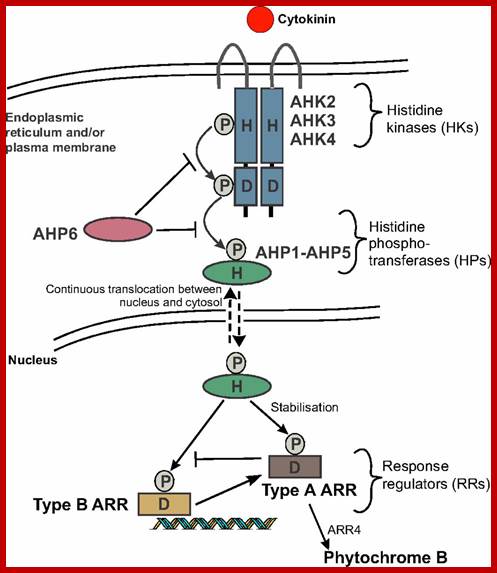
Two component system; http://molbio.mgh.harvard.edu/ www.dev.biologists.org
Cytokinin signalling occurs via a two-component system. The cytokinin receptors Arabidopsis histidine kinases (AHKs) are primarily localized on the endoplasmic reticulum, as well as on the plasma membrane. Cytokinin binds to AHK proteins, inducing conformational changes that trigger a phosphorelay. A phosphoryl group (P) is first transferred from a conserved His (H) to an Asp (D) residue within the receptor and is then relayed to five Arabidopsis histidine phosphotransferase proteins (AHP1-AHP5). The pseudo-HP AHP6 inhibits cytokinin signalling by competing with AHP1-5 for phosphotransfer. The AHPs continuously translocate between the cytosol and the nucleus, where the Arabidopsis response regulators (ARRs) are in turn phosphorylated. Phosphorylation of the type A ARRs stabilizes them. The phosphorylated type B ARRs can bind DNA and initiate transcription of cytokinin-responsive genes, including the type A ARRs, which act as inhibitors of cytokinin signalling. Although type A ARRs are generally considered negative regulators, ARR4 has been shown to upregulate phytochrome B. http://dev.biologists.org
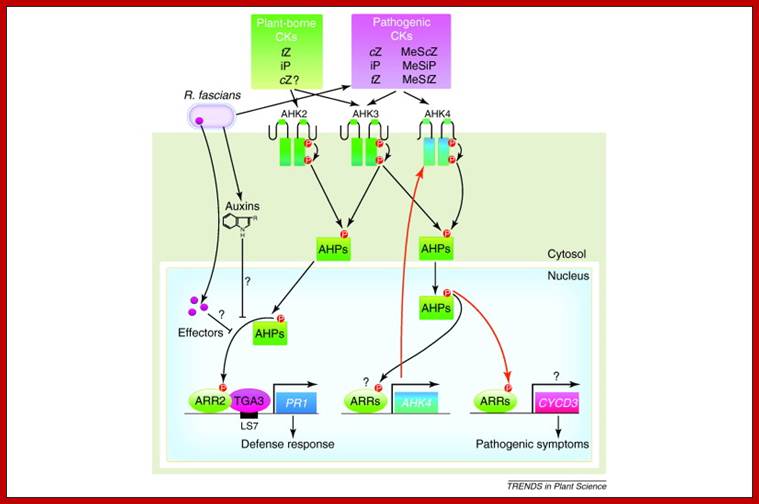
Modulation of plant cytokinin signaling by R. fascians-originated cytokinins. Rhodococcus fascians produces various cytokinin species, including non-degradable 2-methylthio (2-MeS) derivatives of cZ, iP and tZ (MeScZ, MeSiP and MeStZ). Perception of this cytokinin mixture from R. fascians by a cytokinin receptor AHK3 results in abnormal, constitutive activation of cytokinin signaling, including transcriptional induction of AHK4. Accumulation of AHK4 enhances sensitivity to pathogen-derived cytokinins. This results in the de-differentiation of plant cells and leafy gall formation. Although the direct targets of pathogen-derived cytokinins are still elusive, cytokinin-inducible cyclins such as CYCD3 and expansins might trigger de-differentiation as indicated by ectopic induction of KNAT1. It is also possible that R. fascians secretes effectors or auxin to specifically suppress AHK2 and AHK3 – and ARR2-mediated defense responses. Red arrows indicate hyper-activated signaling cascades by R. fascians-derived cytokinins. Abbreviations: AHP, ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN;cZ, cis-zeatin; iP, isopentenyladenine; tZ, trans-zeatin; http://www.cell.com/trends
The major components of the cytokinin signaling pathway were identified in a series of seminal papers around the turn of the century. Cytokinin signaling is mediated by a two-component signaling pathway (Fig. above ) similar to the two-component signaling systems (TCSs) found in bacteria. In brief, cytokinin induces autophosphorylation of a histidine kinase (HK) protein, which results in the transfer of a phosphoryl group from a phospho-accepting histidine residue in the kinase domain to an aspartate residue. The phosphoryl is then transferred to a conserved histidine by histidine phospho transferase (HP) protein. From there, it is finally transferred to an aspartate in the receiver domain of a response regulator (RR). For a thorough overview of cytokinin signaling, the reader is referred to the review by Hwang et al. (Hwang et al., 2012).
Microarray meta-analysis using 13 microarray experiments combined with empirically defined filtering criteria identified a set of 226 genes differentially regulated by cytokinin, a subset of which has previously been validated by other methods. RNA-seq analysis identified 73 cytokinin-regulated genes that were not represented on the ATH1 microarray. study identified several genes that were commonly induced five or repressed twentyeight by cytokinin and auxin in all root three tissues, which could be broadly categorized into metabolic and developmental processes. When compared to auxins, cytokinins repress ~50 or so genes and auxins represss 110 genes. But some genes are upregulated within 2 minutes of treatment and response is insensitive to cycloheximide treatment. A number of related Arabidopsis type-A response regulator genes – ARR3, ARR6,ARR7, ARR8, ARR9, ARR15, and ARR16, which were mostly identified by sequence homology to bacterial response regulators. http://www.ncbi.nlm.nih.gov/
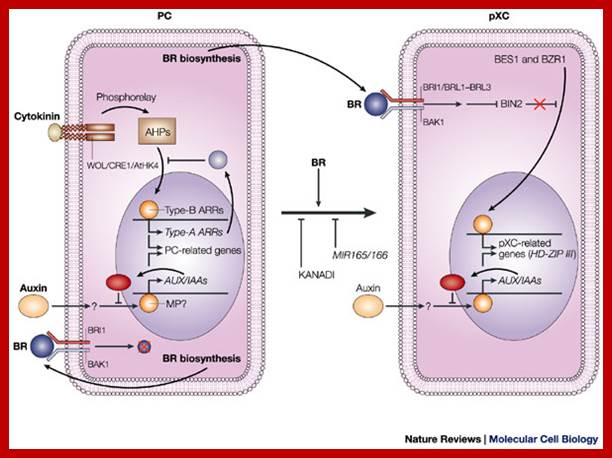
Model for signalling processes in the maintenance and/or the differentiation of procambial cells and xylem cell precursors. Signals that control plant vascular cell differentiation; In procambial cells (PCs), the coordinated signalling by cytokinin and auxin induces the expression of genes that are involved in the maintenance of procambial activities. The auxin-signalling pathway might involve gene expression of auxin-response factors, such as MONOPTEROS (MP), that also function as transcriptional activators, and their repressors, the AUX/IAA proteins. Cytokinin might be perceived by the WOL/CRE1/AtHK4 cytokinin receptor, which, in turn, transmits an intracellular signal that is mediated by a His–Asp phosphorelay mechanism to PC-related histidine-containing phosphotransfer factors (AHPs) and then to PC-related type-B response regulators (ARRs). The type-B ARRs might function as transcriptional activators of PC-related genes including the genes of their repressors, the type-A ARRs. The presence of repressors in auxin- and cytokinin-signalling pathways might allow cytokinin and auxin signalling to be temporal. Brassinosteroids (BRs in the figure) are biosynthesized actively in PCs and secreted, but brassinosteroids do not work as a signal for the maintenance of procambial activities. Instead, brassinosteroids, in the presence of auxin, might initiate differentiation of procambial cells to precursors of xylem cells (pXCs) after recognition by a receptor, which might be a heterodimer composed of either brassinosteroid-insensitive-1 (BRI1) or one of the BRI1-like proteins (BRL1–BRL3), plus BRI1-associated receptor kinase-1 (BAK1). The brassinosteroid signal inactivates the negative regulator BIN2 (brassinosteroid-insensitive-2), which allows the unphosphorylated form of bri1-EMS-suppressor-1 (BES1) and brassinazole-resistant-1 (BZR1) to translocate to the nucleus and to promote pXC-related gene expression. Among the most important pXC-related genes that are induced by brassinosteroids might be the HD-ZIP-III-homeobox gene family, which might function in further xylem cell differentiation. KANADI and the microRNAs MIR165 and MIR166 might suppress differentiation of PCs to pXCs. The suppression by the microRNAs might be caused by the rapid degradation of the HD-ZIP-III gene mRNA through RNAi machinery. Hiroo Fukuda www.nature.com
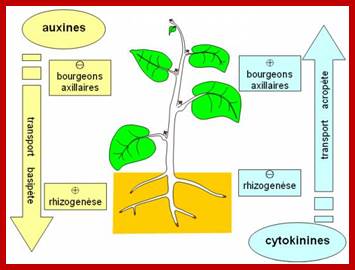
The ratio of auxin to cytokinin plays an important role in the effect of cytokinin on plant growth. Cytokinin alone has no effect on parenchyma cells. When cultured with auxin but no cytokinin, they grow large but do not divide. When cytokinin is added, the cells expand and differentiate. When cytokinin and auxin are present in equal levels, the parenchyma cells form an undifferentiated callus. More cytokinin induces growth of shoot buds, while more auxin induces root formation.[
While cytokinin action in vascular plants is described as pleiotropic, this class of plant hormones specifically induces the transition from apical growth to growth via a three-faced apical cell in moss protonema. This bud induction can be pinpointed todifferentiation of a specific single cell, and thus is a very specific effect of cytokinin
Cytokinin signaling in plants is mediated by a two-component phosphorelay. This pathway is initiated by cytokinin binding to a histidine kinase receptor in the endoplasmic reticulum membrane. This results in the autophosphorylation of the receptor, with the phosphate then being transferred to a phosphotransfer protein. The phosphotransfer proteins can then phosphorylate the type-B response regulators (RR) which are a family of transcriptions factors. The phosphorylated, and thus activated, type-B RRs regulate the transcription of numerous genes, including the type-A RRs. The type-A RRs negatively regulate the pathway.
Auxin is known to regulate the biosynthesis of cytokinin. But Cytokinins have recently been found to play a role in plant pathogenesis. For example, cytokinins have been described to induce resistance against Pseudomonas syringae inArabidopsis thaliana[14] and Nicotiana tabacum.[15] Also in context of biological control of plant diseases cytokinins seem to have potential functions. Production of cytokinins byPseudomonas fluorescens G20-18 has been identified as a key determinant to efficiently control the infection of A. thaliana with P. syringae; https://en.wikipedia.org
In procambial cells (PCs), the coordinated signaling by cytokinin and auxin induces the expression of genes that are involved in the maintenance and growth of procambial activities into xylem elements. The auxin-signaling pathway might be involved in gene expression of auxin-response factors, such as MONOPTEROS (MP), that also function as transcriptional activators, and their repressors, the AUX/IAA proteins. Cytokinin might be perceived by the WOL/CRE1/AtHK4 cytokinin receptor, which, in turn, transmits an intracellular signal that is mediated by a His–Asp phosphorelay mechanism to PC-related histidine-containing phosphotransfer factors (AHPs) and then to PC-related type-B response regulators (ARRs). The type-B ARRs might function as transcriptional activators of PC-related genes including the genes of their repressors, the type-A ARRs. The presence of repressors in auxin- and cytokinin-signaling pathways might allow cytokinin and auxin signaling to be temporal. Brassinosteroids (BRs in the figure) are biosynthesized actively in PCs and secreted, but brassinosteroids do not work as a signal for the maintenance of procambial activities. Instead, brassinosteroids, in the presence of auxin, might initiate differentiation of procambial cells to precursors of xylem cells (pXCs) after recognition by a receptor, which might be a heterodimers, composed of either brassinosteroid-insensitive-1 (BRI1) or one of the BRI1-like proteins (BRL1–BRL3), plus BRI1-associated receptor kinase-1 (BAK1). The brassinosteroid signal inactivates the negative regulator BIN2 (brassinosteroid-insensitive-2), which allows the unphosphorylated form of bri1-EMS-suppressor-1 (BES1) and brassinazole-resistant-1 (BZR1) to translocate to the nucleus and to promote pXC-related gene expression. Among the most important pXC-related genes that are induced by brassinosteroids might be the HD-ZIP-III-homeobox gene family, which might function in further xylem cell differentiation. KANADI and the microRNAs MIR165 and MIR166 might suppress differentiation of PCs to pXCs. The suppression by the microRNAs might be caused by the rapid degradation of the HD-ZIP-III gene mRNA through RNAi machinery.
CRF facilititates Calcium release.http://www.cell.com
The above diagram shows how cytokinins play a role in localization of CRF6 in cytosol.
Regulation of cytokinin biosynthesis, compartmentalization and translocation:
Response Factors in Arabidopsis :
Cytokinins induce CRF genes. There are 23 such CRFs; they are cytokinin response/regulatory (CRFs) factors and transcriptional activators. Aclosely related six of them rlated to APETALA2 (AP2) Are transcriptional factors. They play important role in plant development. Mutants in CRF genes produce malfunctions in leaf and cotyledon development as shown in the diagrams above. They are divided into three groups, two of which A and B are involved into cytokinin signaling. Phosphorylation of A stabilizes them where as phosphorylation of B ARR enables them to bind to DNA specific promoter or regulatory sequences and initiate transcription, (Rashotte et al 2006). One prominent class of genes that has received special attention includes those that are involved in crosstalk with other hormonal pathways; these genes, which include signalling genes responsive to auxin (SHY2/IAA3, AXR3/IAA17) and gibberellins (GNL/CGA1/GATA22, GNC/GATA21), have recently been reviewed by Brenner et al. (Brenner et al., 2012).
Recent studies show that cytokinins cross talks with other hormones, especially Auxins. Cross talks between cytokinins with other has been studied , but interaction with Auxin is very well illustrated and known.
http://dev.biologists.org/
Cytokinin interaction is well understood (not completely) at shoot apex, axillary bud, at lateral root initiation zone, root transcription zone and root meristem; auxins upregulate the synthesis of Cytokinins.
function
in plant growth and d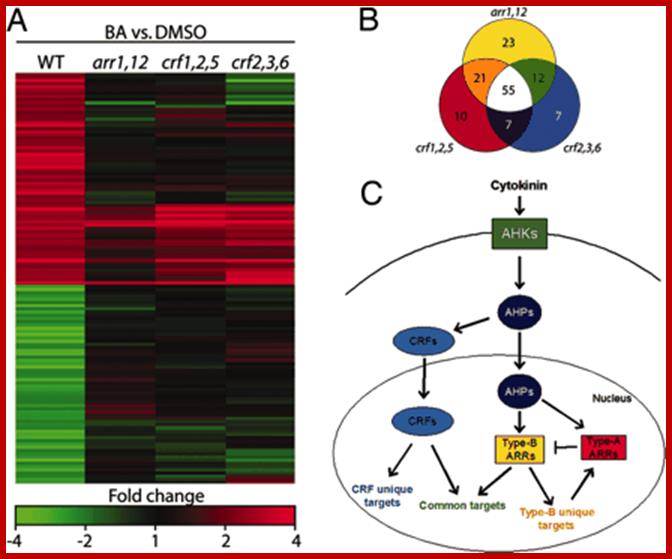
Cytokinin activated gene expression is shown in the form of microarray. http://www.cell.com
CRFs act in parallel with type-B ARRs to mediate cytokinin regulated gene expression. (A) Wild-type, arr1,12, crf1,2,5, and crf2,3,6 seedlings were treated with either 10 μM BA or a DMSO control for 1 h and gene expression analyzed by using a microarray. Genes that displayed a ≥2-fold change in response to cytokinin in the wild type are shown. (B) Venn diagram of the 135 cytokinin-regulated genes affected by the arr1,12, crf1,2,5, and/or crf2,3,6 mutations. (C) Model of cytokinin signaling. Both AHPs and CRFs move into the nucleus in response to cytokinin. Once there, the AHPs phosphorylate the type-B ARRs, which, together with CRFs, mediate cytokinin-regulated gene expression. Another diagram shows few more Cytokinin signaling features.www.pnas.org
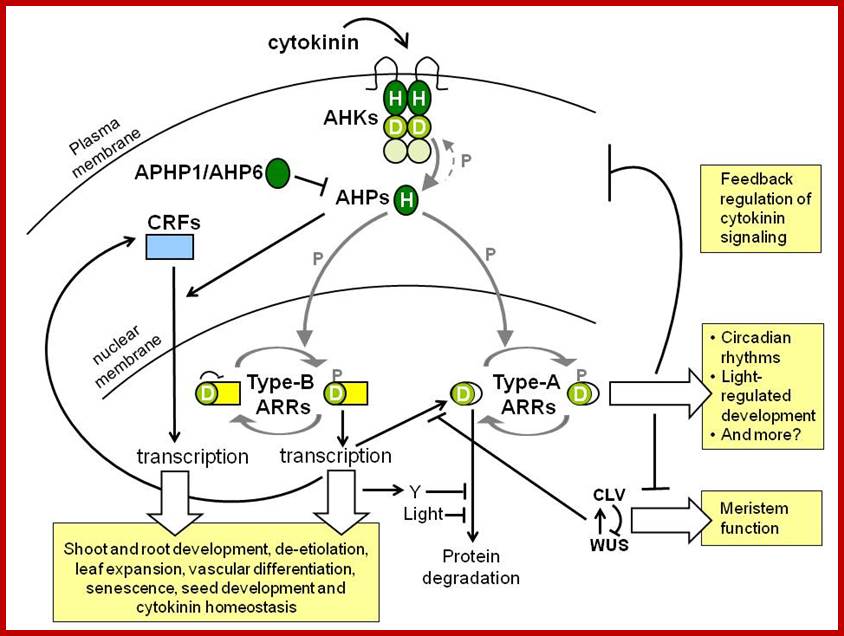
Crossing Path: cytokinin signalling and cross talk:
Cytokinins are a major class of plant hormones that are involved in various aspects of plant development, ranging from organ formation and apical dominance to leaf senescence. Cytokinin and auxin have long been known to interact antagonistically, and more recent studies have shown that cytokinins also interact with other plant hormones to regulate plant development. A growing body of research has begun to elucidate the molecular and genetic underpinnings of this extensive crosstalk. The rich interconnections between the synthesis, perception and transport networks of these plant hormones provide a wide range of opportunities for them to modulate, amplify or buffer one another.
Cytokinins signalling take place like bacterial two component system. Cytokinins receptors in Arabidopsis Histidine Kinase AHK are located on ER and Plasma membranes. Cytokinin binding to AHK induces phosphorelay, where phospho group is transferred from His (H) to Asp (D) within the receptor, then relayed to five AHP 1-5. The AHP6 (pseudo AHP0 inhibits cytokinins signalling. AHPs constantly translocate between cytosol and the Nucleus. In the nucleus ARRs (Arabidopsis receptor regulators in turn are phosphorylated. P-lated ARR-B binds to DNA promoter elements of cytokinins responsive genes including type A ARRs Inhibitory, but ARR4 up regulates Phytochrome B.
There is extensive literature regarding regulation of the cell cycle in yeast and animal cells (reviewed in [6,7]). Cell cycle progression is controlled at the G1→S and G2→M checkpoints, primarily by two classes of proteins: cyclins, and cyclin dependent kinases (CDKs) (Figure 1). Passage through the checkpoints requires the activation of CDKs, and this is achieved by association with cyclins and by altering the phosphorylation state of the CDK [8,9]. By associating with different cyclins, cdc2, the first CDK identified, controls both G1→S and G2→M transition in yeast. Animal cells have families of CDKs, some similar to cdc2, that act at G2→M, and others that are distinct and act exclusively at G1→S. B-type cyclins are the major class of mitotic cyclins, which act at the G2→M transition, and D-type cyclins are the major class involved in G1→S transition. cDNAs encoding CDKs and G1 and mitotic cyclins have been isolated in plants, and CDK inhibitors have been shown to block cell cycle progression at both G1→S and G2→M in Arabidopsis and Petunia cells [10–12]. Therefore, it is likely that these proteins also mediate the cell cycle in plants. Given the link between cytokinins and cell division, a natural question that arises is whether cytokinins affect the expression or activity of these cell cycle regulatory proteins. Several observations suggest that cytokinins may play a role in the G2→M transition.
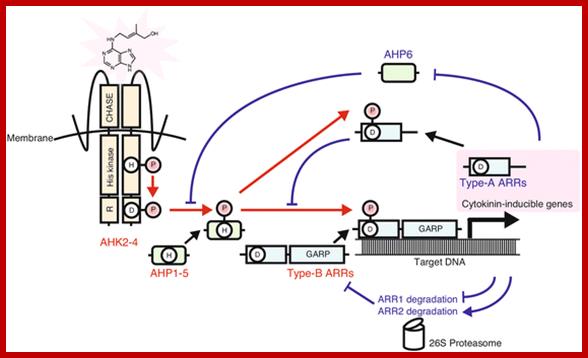
Diagram of the cytokinin two component system (TCS). AHKs (AHK2, AHK3 and AHK4/WOL1/CRE1) are autophosphorylated in response to cytokinins. The phosphoryl group is transferred to type-B ARRs through AHPs. Phosphorylated type-B ARRs bind to target DNA and induce the expression of a set of genes involved in cytokinin primary response. The stability of type-A ARRs, which repress cytokinin TCS signaling, is controlled through proteolysis by the 26S proteasome in a feedback loop. Expression of AHP6, which inhibits phosphotransfer between AHKs and canonical AHPs, is repressed by cytokinin. Red arrowsindicate phosphotransfer. Blue solid arrows and T-end lines represent positive and negative regulation, respectively; http://bmcbiol.biomedcentral.com/
Cytokinins transduction leads to activation of certain genes, they use cytokinin response motif CRM for BARR proteins [AGAT] T,C as cytokinin response motifs. AAGAT[T,C]TT for ARR1 cytokinin response motifs –ECRM. There are five more motifs cytokinins response elements in cytokinin response promoters; CATATATA, TATATATA, TATATTCC, TATATTTA, and TGTATTTC, albeit some with a p-value between 0.05 and 0.06.

The genome-wide transcriptional response of the
model organism Arabidopsis thaliana to cytokinin has been investigated by
different research groups as soon as large-scale transcriptomic techniques
became affordable. Over the last 10 years many transcriptomic datasets related
to cytokinin have been generated using different technological platforms, some
of which are published only in databases, culminating in an RNA sequencing
experiment. Two approaches have been made to establish a core set of
cytokinin-regulated transcripts by meta-analysis of these datasets using
different preferences regarding their selection. Here we add another meta-analysis
derived from an independent microarray platform (CATMA), combine all the
meta-analyses available with RNAseq data in order to establish an advanced core
set of cytokinin-regulated transcripts, and compare the results with the
regulation of orthologous rice genes by cytokinin. We discuss the functions of
some of the less known cytokinin-regulated genes indicating areas deserving
further research to explore cytokinin function. Finally, we investigate the
promoters of the core set of cytokinin-induced genes for the abundance and
distribution of known cytokinin-responsive cis elements and identify a set of
novel candidate motifs.
Summarizing and exploring data of a decade of cytokinin-related
transcriptomics (PDF Download Available). Available from: https://www.researchgate.net/publication/_cytokinin-related_transcriptomics
[accessed Jun 15, 2017].;CRFs;
Cytokinins response elements in cytokinin response promoters; Wolfram G Brenner and Thomas Schmülling; https://www.researchgate.net/
Sequence logo of a novel model of a potential type –B response regulator binding site, 5′-GAT(T/C)-3′; The motif combines al single motifs found in the analysis pictured in the figure, promoter elements of of cytokinin-regulated genes. http://journal.frontiersin.org/

The rice cytokinin signaling pathway is most likley similar to that of Arabidopsis. We the authors of the above diagram have focused on the disruption of the OsAHPs, which are positive elements represented by only two genes in the rice genome, and the type-A OsRRs, which, at least in Arabidopsis are negative regulators of cytokinin signaling. http://labs.bio.unc.edu/
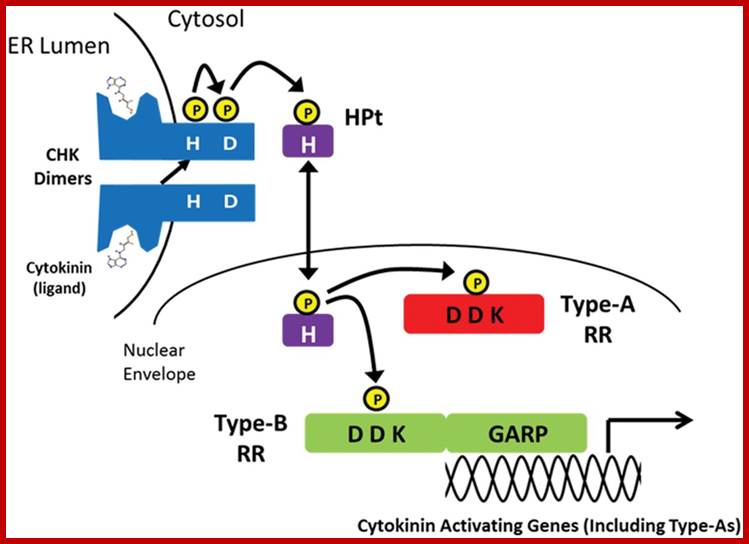
Cytokinin is an essential plant hormone that is involved in a wide range of plant growth and developmental processes which are controlled through its signalling pathway. Cytokinins are a class of molecules that are N6-substituted adenine derivatives, such as isopentenyl adenine, and trans- and cis-zeatin, which are common in most plants. The ability to perceive and respond to cytokinin occurs through a modified bacterial two-component pathway that functions via a multi-step phosphorelay. This cytokinin signalling process is a crucial part of almost all stages of plant life, from embryo patterning to apical meristem regulation, organ development and eventually senescence. The cytokinin signalling pathway involves the co-ordination of three types of proteins: histidine kinase receptors to perceive the signal, histidine phosphotransfer proteins to relay the signal, and response regulators to provide signal output. This pathway contains both positive and negative elements that function in a complex co-ordinated manner to control cytokinin-regulated plant responses. Although much is known about how this cytokinin signal is perceived and initially regulated, there are still many avenues that need to be explored before the role of cytokinin in the control of plant processes is fully understood. Erika A. Keshishian, Aaron M. Rashotte http://essays.biochemistry.org
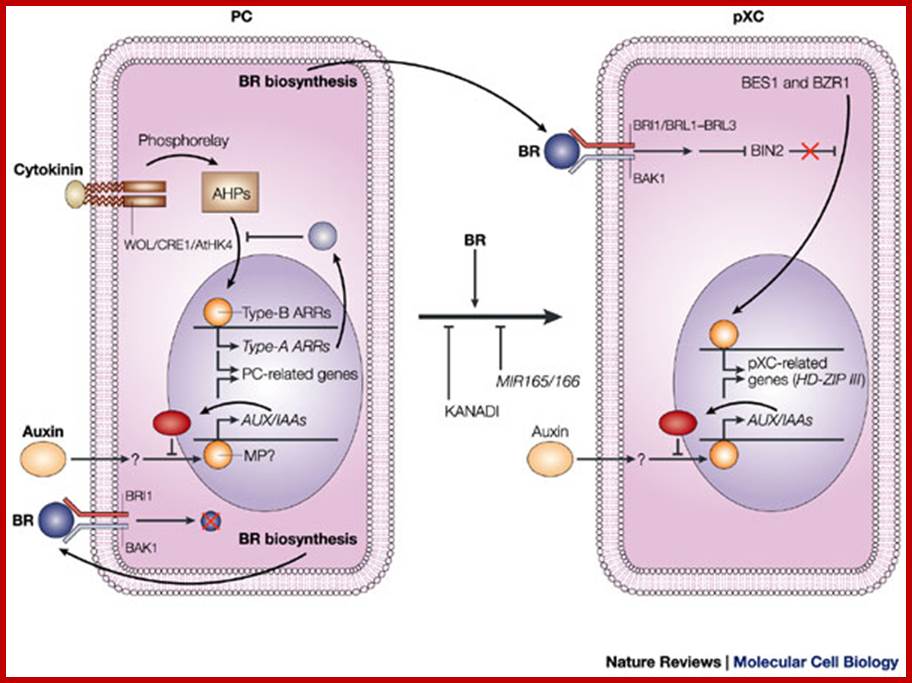
Model for signalling processes in the maintenance and/or the differentiation of procambial cells and xylem cell precursors. Signals that control plant vascular cell differentiation; In procambial cells (PCs), the coordinated signalling by cytokinin and auxin induces the expression of genes that are involved in the maintenance of procambial activities. The auxin-signalling pathway might involve gene expression of auxin-response factors, such as MONOPTEROS (MP), that also function as transcriptional activators, and their repressors, the AUX/IAA proteins. Cytokinin might be perceived by the WOL/CRE1/AtHK4 cytokinin receptor, which, in turn, transmits an intracellular signal that is mediated by a His–Asp phosphorelay mechanism to PC-related histidine-containing phosphotransfer factors (AHPs) and then to PC-related type-B response regulators (ARRs). The type-B ARRs might function as transcriptional activators of PC-related genes including the genes of their repressors, the type-A ARRs. The presence of repressors in auxin- and cytokinin-signalling pathways might allow cytokinin and auxin signalling to be temporal. Brassinosteroids (BRs in the figure) are biosynthesized actively in PCs and secreted, but brassinosteroids do not work as a signal for the maintenance of procambial activities. Instead, brassinosteroids, in the presence of auxin, might initiate differentiation of procambial cells to precursors of xylem cells (pXCs) after recognition by a receptor, which might be a heterodimer composed of either brassinosteroid-insensitive-1 (BRI1) or one of the BRI1-like proteins (BRL1–BRL3), plus BRI1-associated receptor kinase-1 (BAK1). The brassinosteroid signal inactivates the negative regulator BIN2 (brassinosteroid-insensitive-2), which allows the unphosphorylated form of bri1-EMS-suppressor-1 (BES1) and brassinazole-resistant-1 (BZR1) to translocate to the nucleus and to promote pXC-related gene expression. Among the most important pXC-related genes that are induced by brassinosteroids might be the HD-ZIP-III-homeobox gene family, which might function in further xylem cell differentiation. KANADI and the microRNAs MIR165 and MIR166 might suppress differentiation of PCs to pXCs. The suppression by the microRNAs might be caused by the rapid degradation of the HD-ZIP-III gene mRNA through RNAi machinery. Hiroo Fukuda; http://www.nature.com/
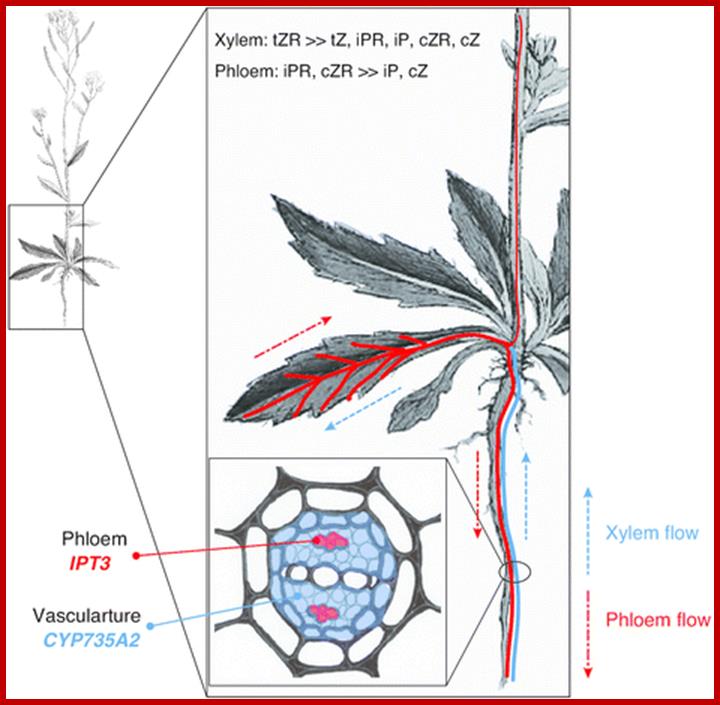
Spatial expression patterns of IPT3 and CYP735A2 in Arabidopsis. Spatial expression patterns of IPT3 and CYP735A2 are indicated in red and blue, respectively. IPT3 is predominantly expressed in phloem, and CYP735A2 in root vasculature. tZR is the major form of xylem cytokinins, and iPR and cZR are found in phloem [40], suggesting that iP-type and tZ-type cytokinins are directionally translocated between organs. The importance of cytokinins’ local action is best characterized by their role in maintenance of shoot apical meristem and root vascular development.
The Arabidopsis picture is modified from Sowerby et al
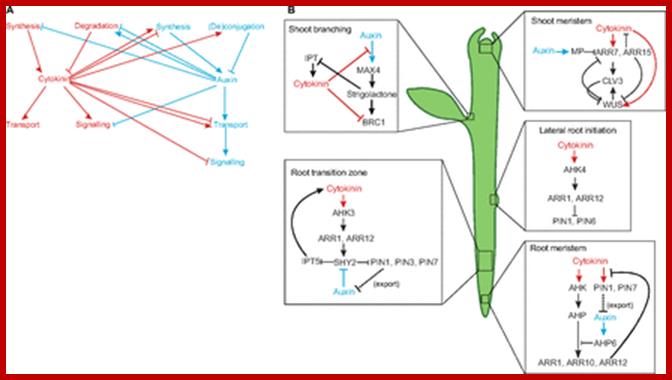
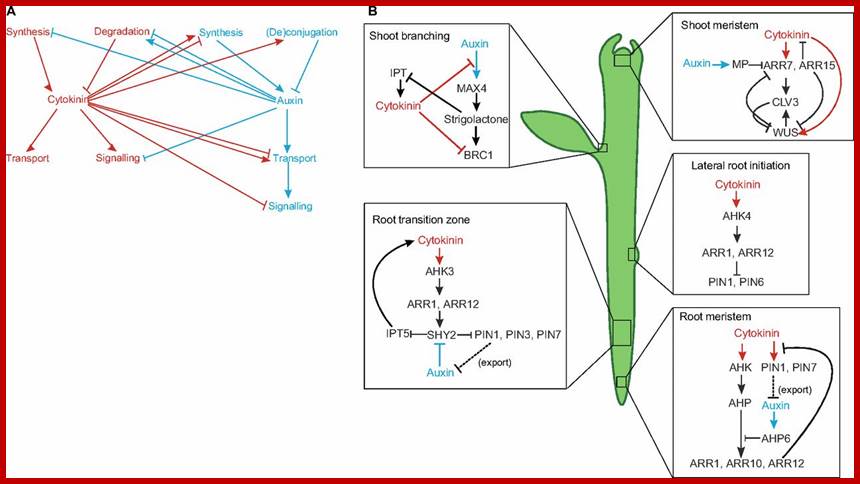
Cytokinin interacts with auxin in several developmental contexts. (A) There are multiple points of interaction between the auxin (blue) and cytokinin (red) pathways. In addition to the inter-hormonal interactions shown, both hormones regulate their own metabolism and perception, adding further complexity to the pathways. (B) Interactions between auxin and cytokinin are crucial for many important aspects of plant development (e.g. in shoot branching, the shoot meristem, the root transition zone, lateral root initiation and in the root meristem; see main text for details) and most of these interactions involve mutual feedback loops. Auxin and auxin activity are depicted in blue; cytokinin and cytokinin activity are depicted in red. AHK, Arabidopsis histidine kinase 4; AHP, Arabidopsis histidine phosphotransferase; ARR, Arabidopsis response regulator; BRC1, branched 1; CLV3, clavata 3; IPT, isopentenyltransferase; MAX4, more axillary branching 4; MP, monopteros; PIN, pin-formed; SHY, short hypocotyl; WUS, wuschel; dorgev.biologists. http://dev.biologists.org/
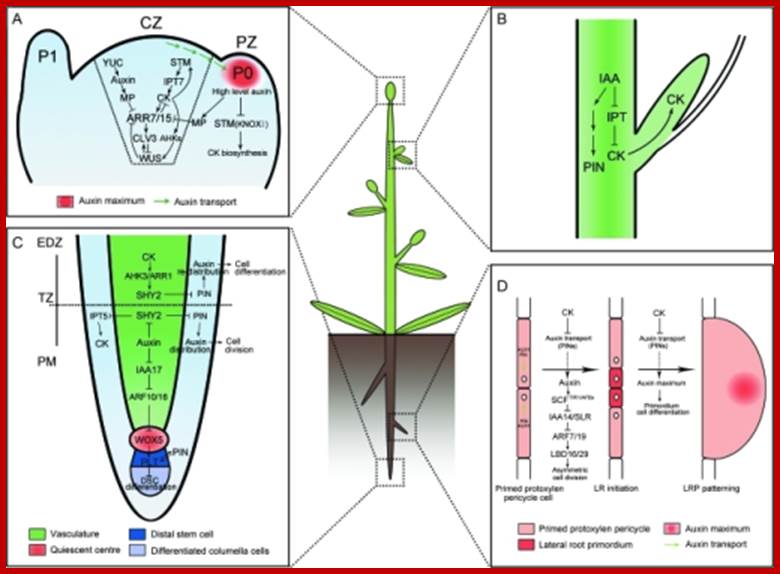
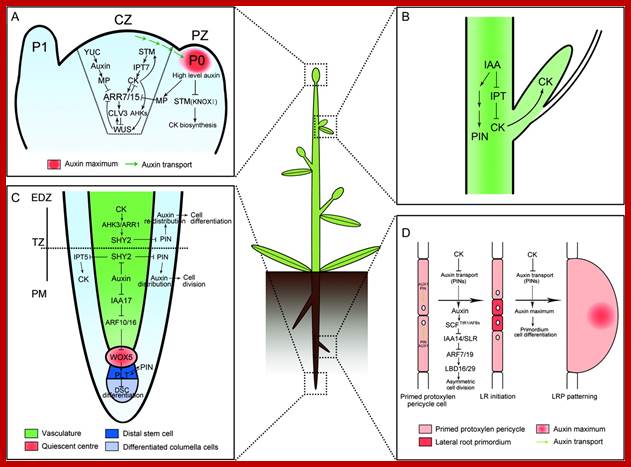
Molecular Mechanisms of Auxin and Cytokinin Interaction in the Regulation of Plant Meristem Development.(A) In CZ of shoot meristem, ARR7 and ARR15 act as integrative factors in auxin and cytokinin signaling pathways. Auxin represses the expression of ARR7 and ARR15 while cytokinin promotes their expression through a STM-dependent pathway. Both of them regulate the expression of WUS in a negative feedback loop, critical for stem-cell formation. During the formation of lateral organ primordia, a high level of auxin transported from CZ blocks the biosynthesis of cytokinin by suppressing KNOXI function in PZ. CZ, central zone; PZ, peripheral zone; P0/P1, organ primordia.(B) Auxin is transported from the shoot apex to repress cytokinin biosynthesis, leading to the inhibition of axillary bud growth.(C) In the root meristem, auxin promotes the expression of PINs through the degradation of SHY2 proteins, resulting in the maintenance of an auxin gradients and cell division. In contrast, cytokinin impedes the expression of PINs by stimulating the expression of SHY2, leading to auxin redistribution and cell differentiation. Auxin also plays an important role in the differentiation of root DSC by mediating the expression of WOX5 and PLT. PM, proximal meristem; EDZ, elongation differentiation zone; TZ, transition zone; DSC, distal stem cell.(D) In certain xylem pole pericycle cells, the transport and perception of auxin trigger an asymmetric cell division critical for the LR initiation and LRP patterning. By contrast, cytokinin negatively regulates the LR initiation and LRP patterning by inhibiting the expression of PINs and the auxin distribution gradients. LR, lateral roots; LRP, lateral root primordial; http://openi.nlm.nih.gov/
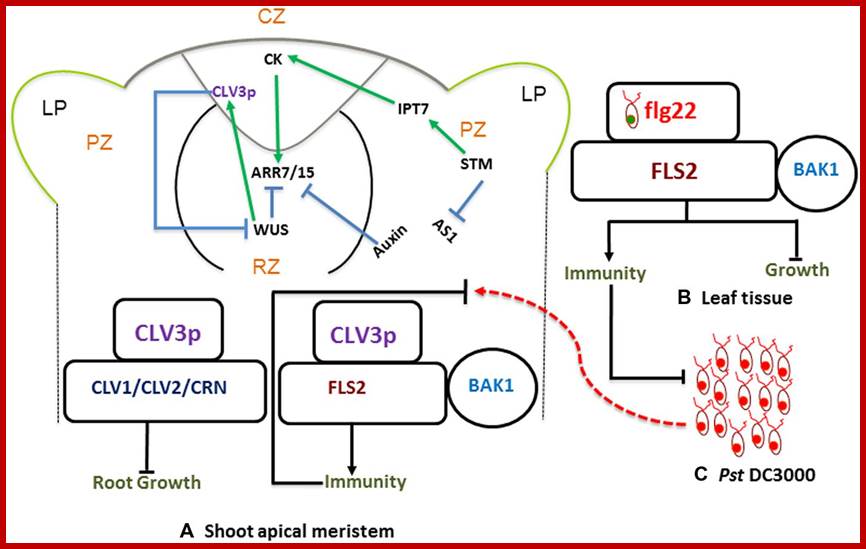
Stem Cells Triggered Immunity; Schematic diagram showing shoot apical meristem (A) protective and regulatory networks, driven by auxin and cytokinins (CKs). WUS and CLV3p mediated regulation play a pivotal role in maintaining a balance between proliferation and differentiation. CLV3p is perceived by the CLV-receptor complex and leads to maintenance of stem cell (left). There is an independent pathway where CLV3 also binds to FLS2 and activates innate immunity (right). This is analogous to typical bacterial flagellin perception mechanism, where flg22-FLS2 interaction triggers innate immunity in the leaf tissue (B) against infection with bacterial pathogens such as Pst DC3000 (C). http://journal.frontiersin.org/
Stem-cell-triggered immunity safeguards cytokinin enriched plant shoot apexes from pathogen infection:
Intricate mechanisms discriminate between friends and foes in plants. Plant organs deploy overlapping and distinct protection strategies. Despite vulnerability to a plethora of pathogens, the growing tips of plants grow bacteria free. The shoot apical meristem (SAM) is among three stem cells niches, a self-renewable reservoir for the future organogenesis of leaf, stem, and flowers. How plants safeguard this high value growth target from infections was not known until now. Recent reports find the stem cell secreted 12-amino acid peptide CLV3p (CLAVATA3 peptide) is perceived by FLS2 (FLAGELLIN SENSING 2) receptor and activates the transcription of immunity and defense marker genes. No infection in the SAM of wild type plants and bacterial infection in clv3 and fls2 mutants illustrate this natural protection against infections. Cytokinins (CKs) are enriched in the SAM and regulate meristem activities by their involvement in stem cell signaling networks. Auxin mediates plant susceptibility to pathogen infections while CKs boost planpot immunity. Here, in addition to the stem-cell-triggered immunity we also highlight a potential link between CK signaling and CLV3p mediated immune response in the SAM. Muhammad Naseem†† and ; Mugdha Srivatsav and Thomas Dandekar ; http://journal.frotiersin.org,
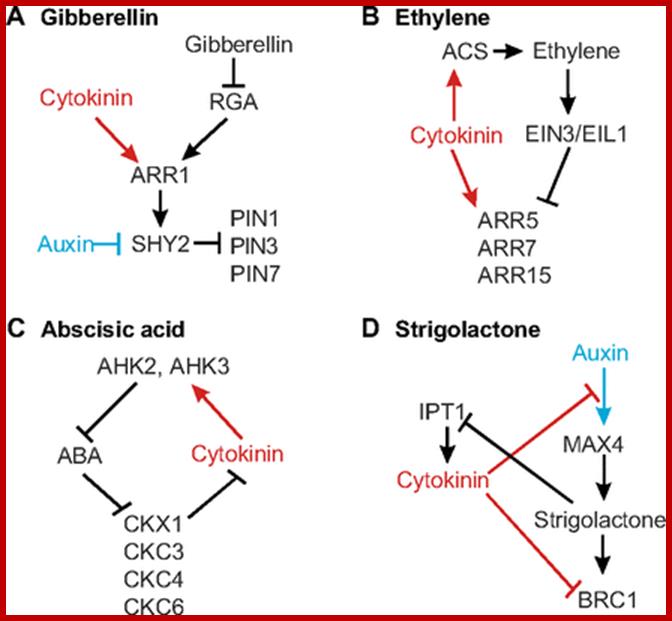
Interactions between cytokinin and other hormones. (A) Cytokinin and gibberellin signalling come together in the regulation of ARR1, the expression of which is repressed by gibberellin (via degradation of the DELLA protein RGA, which promotes ARR1 expression) and promoted by cytokinin. The regulation ofSHY2 by ARR1 also represents a point of crosstalk with auxin, thus connecting three hormones in one network. (B) Ethylene and cytokinin signalling converge in the control of the type A ARRs ARR5, ARR7 and ARR15, which play a role in freezing tolerance. Cytokinin also increases ethylene biosynthesis by increasing the stability of ACS proteins, which catalyse the rate-limiting step in ethylene biosynthesis. (C) Components of the cytokinin two-component signalling system interact with abscisic acid (ABA), regulating salinity and drought response. ABA represses several CKX genes, whereas AHK2 and AHK3 downregulate many ABA-responsive genes. The loop shown in the figure will buffer against changes in the concentration of either hormone, though the interaction is likely to be more complex in reality. (D) Cytokinin interacts with strigolactones to regulate the outgrowth of axillary buds. In addition to inhibiting the transcription factor BRC1, which acts downstream of strigolactones, cytokinins prevent auxin-mediated regulation of the strigolactone biosynthesis gene MAX4. Strigolactones, in turn, negatively regulate cytokinin biosynthesis through IPT1. Auxin and auxin activity are depicted in blue; cytokinin and cytokinin activity are depicted in red. ACS, aminocyclopropane-1-carboxylic acid synthase; AHK, Arabidopsis histidine kinase 4; ARR, Arabidopsis response regulator; BRC1, branched 1; EIL1, ethylene-insensitive 3-like 1; EIN3, ethylene-insensitive 3; IPT1, isopentenyltransferase 1; MAX4, more axillary branching; PIN, pin-formed; SHY, short hypocotyl. http://dev.biologists.org/
Callus:
These data collectively suggest that CIM induces callus through the genetic pathway mediating lateral root initiation and that CIM-induced callus, at least in Arabidopsis, is not as dedifferentiated as previously thought.
Auxin and cytokinin have been widely used to generate callus, but surprisingly little is known about how they induce callus at the molecular level. Several recent studies demonstrated that various regulators of lateral root development participate in callus formation on CIM.
Fan et al. (2012) have shown that the expression of LBD16, LBD17, LBD18, and LBD29 ,Lateral organ Boundaries Domain, is up regulated by CIM, Callus Inducing Medium and that overexpression of each of the four is sufficient to induce callus with a similar appearance to CIM-induced callus.
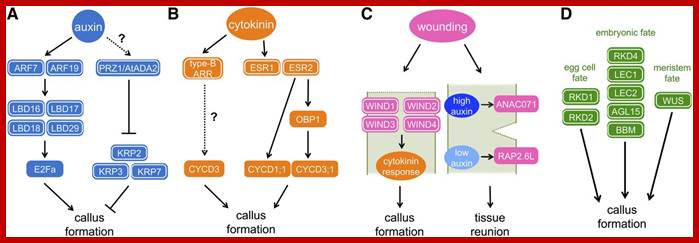
http://www.plantcell.org/;http://www.plantcell.org/
Molecular Mechanisms of Callus Induction.
(A) Auxin-induced callus formation. Auxin signaling is transduced via ARF transcription factors, especially ARF7 and ARF19, to activate the expression of LBD family transcription factors, LBD16, LBD17, LBD18, and LBD29. These LBDs in turn induce E2Fa, a transcription factor that plays a central role in cell cycle reentry. The PRZ1/AtADA2 protein mediates auxin-dependent repression of CDK inhibitors, KRP2, KRP3, and KRP7. How auxin modulates the expression and/or activity of PRZ1/AtADA2 is currently unknown.
(B) Cytokinin-induced callus formation. Cytokinin signaling is transduced via two-component regulatory pathway to activate the type-B ARR transcription factors. The expression of CYCD3;1 is sharply upregulated by cytokinin, but whether it is directly activated by type-B ARR is not known. The AP2/ERF transcription factor ESR1 is also upregulated by cytokinin. ESR1 and its functionally redundant homolog ESR2 might mediate cell cycle reactivation since ESR2 induces the expression of CYCD1;1 as well as a DOF binding transcription factor OBP1. OBP1 is thought to promote the cell cycle progression by inducing expression of CYCD3;3 and several other cell cycle regulators.
(C) Wound-induced callus formation. Complete excision of the Arabidopsishypocotyls induces the expression of WIND1, WIND2, WIND3, and WIND4genes at the wound site, which in turn upregulates the cytokinin response to promote callus formation. When Arabidopsis stems are half-cut, auxin transported from the shoot apex accumulates at the upper end of the wound site, which then induces the expression of ANAC071 gene. Auxin is depleted from the lower end, resulting in the induction of the RAP2.6L gene. Both of these responses are required for the local activation of cell proliferation to heal the gap at the wound site. Dotted lines indicate the wound site.
(D) Callus formation by the reacquisition of embryonic or meristematic fate. Overexpression of each of the master regulators in the egg cell fate (RKD1 and RKD2), embryonic fate (RKD4, LEC1, LEC2, AGL15, and BBM), or meristem fate (WUS) is sufficient to induce callus formation. Proteins with confirmed function in callus formation are highlighted with white circles, while those inferred in callus formation based on indirect evidence are unmarked.
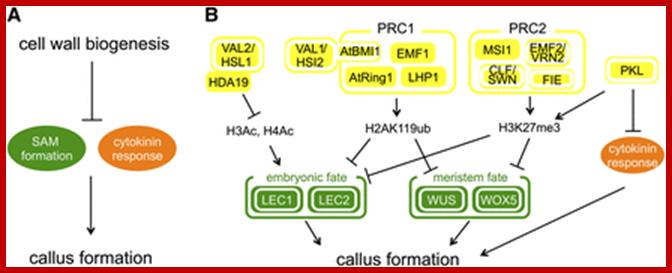
http://www.plantcell.org/;http://www.plantcell.org/
Molecular Mechanisms of Callus Repression.
(A) Orderly deposition of cell wall polysaccharides prevents ectopic callus formation. Defects in cell wall biosynthetic enzymes (e.g., nolac-H18 in tobacco and tsd1 and tsd2 in Arabidopsis) result in the ectopic expression of shoot apical meristem (SAM) genes and increased cytokinin response, leading to callus induction as an indirect downstream consequence.
(B) Ectopic callus formation is repressed by multiple epigenetic mechanisms. The histone deacetylase HDA19 interacts with VAL2/HSL1 to repress the expression of embryonic regulators, such as LEC1 and LEC2 via deacetylation of histone H3 (H3Ac) and H4 (H4Ac). The Polycomb group proteins, PRC1 and PRC2, repress the expression of both embryonic and meristematic regulators (WUS, WOX5, and others) through monoubiquitination of H2A at Lys-119 (H2AK119ub) and trimethylation of histone H3 at Lys-27 (H3K27me3), respectively. The VAL1/HSI2 protein physically interacts with At BMI1 and may recruit PRC1 to target loci for their repression. The CHD3/4-like chromatin remodeling protein PKL participates in the deposition of H3K27me3 on the Polycomb targets. In addition, PKL may repress cytokinin response through histone deacetylation. Proteins with confirmed function in callus formation are highlighted with white circles, while those inferred in callus formation based on indirect evidence are unmarked.
Callus:
Plants develop unorganized cell masses like callus and tumors in response to various biotic and abiotic stimuli. Since the historical discovery that the combination of two growth-promoting hormones, auxin and cytokinin, induces callus from plant explants in vitro, this experimental system has been used extensively in both basic research and horticultural applications. The molecular basis of callus formation has long been obscure, but we are finally beginning to understand how unscheduled cell proliferation is suppressed during normal plant development and how genetic and environmental cues override these repressions to induce callus formation. In this review, we will first provide a brief overview of callus development in nature and in vitro and then describe our current knowledge of genetic and epigenetic mechanisms underlying callus formation. callus= callum (latin); Under certain conditions, callus cells also undergo somatic embryogenesis, a process in which embryos are generated from adult somatic cells (Steward et al., 1958). Thus, at least some forms of callus formation are thought to involve cell dedifferentiation. However, it has also been acknowledged that calli are very diverse and can be classified into subgroups based on their macroscopic characteristics. For example, calli with no apparent organ regeneration typically are called friable or compact callus (Figure 1A). Other calli that display some degrees of organ regeneration are called rooty, shooty, or embryonic callus, depending on the organs they generate (Zimmerman, 1993;Frank et al., 2000) (Figure 1A). It is also known that different types of callus inArabidopsis thaliana have distinct gene expression profiles (Iwase et al., 2011a). Therefore, the term callus includes cells with various degrees of differentiation.
We will then summarize our current knowledge of how plants reprogram their differentiation status and regain proliferative competence to produce callus. Finally, we will describe genetic and epigenetic mechanisms that repress callus induction during postembryonic development in plants.
Generally speaking, an intermediate ratio of auxin and cytokinin promotes callus induction, while a high ratio of auxin-to-cytokinin or cytokinin-to-auxin induces root and shoot regeneration, respectively ;
callus-inducing medium (CIM) ; these calli are not a mass of unorganized cells; instead, they have organized structures resembling the primordia of lateral roots http://www.plantcell.org/;http://www.plantcell.org/
http://slideplayer.com