PHYSIOLOGY OF FLOWERING
Plants, to begin with go through a period of vegetative growth. The extent of vegetative growth is endowed with its genetic potentiality. Accordingly, they may grow into herbs or shrubs and some may develop into trees or climbers. Generally, every plant after going through a period of vegetative growth, responding to environmental clues, start producing floral structures, which may be in the form of characteristic single flowers or inflorescences.

Nelumbium flower;www.flickrhivemind.net

Onagraceae-Evenig primrose member; http://www.wildflowers-and-weeds.com/
Many plants for that matter, a large number of plant species (higher plants), after a period of vegetative growth, start flowering irrespective of the season. But some plants flower only in a particular season of the year. Based on the duration of vegetative growth required for the plants to produce flowers, they have been classified into annuals, biennials and perennials. All plants have to acquire ripeness to flowering. Annuals complete their vegetative growth and flowering in one season and then they die. Biennials produce vegetative growth in one season and flower in the next season and die. But perennials remain for many years and flower seasonally.

Nelumbo nucifera; Lotus flower; www.flickr.com

Catharanthus roseus; https://en.wikipedia.org; Alabama plants.com
In fact, some trees do not flower till they reach a certain age. For example, coconut and areca- nut plants start producing flowers only when they reach an age of 6-8 years. On the other hand in the case of bamboo plants, they grow for a number of years, and flower only once in their life span. As soon as they flower, produce seeds and plants die (monocarpic plants). Interestingly there are many plants which flower throughout the year, ex. Catharathus roseus called Nithya mallige in Kannada; Apocyanaceae.

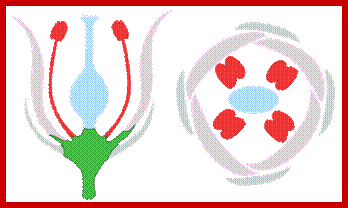
Parts of a flower from outer to the central region; http://biology.tutorvista.com/

Aestivation; www.biology.tutorvista.com

http://www.mun.ca/Biology
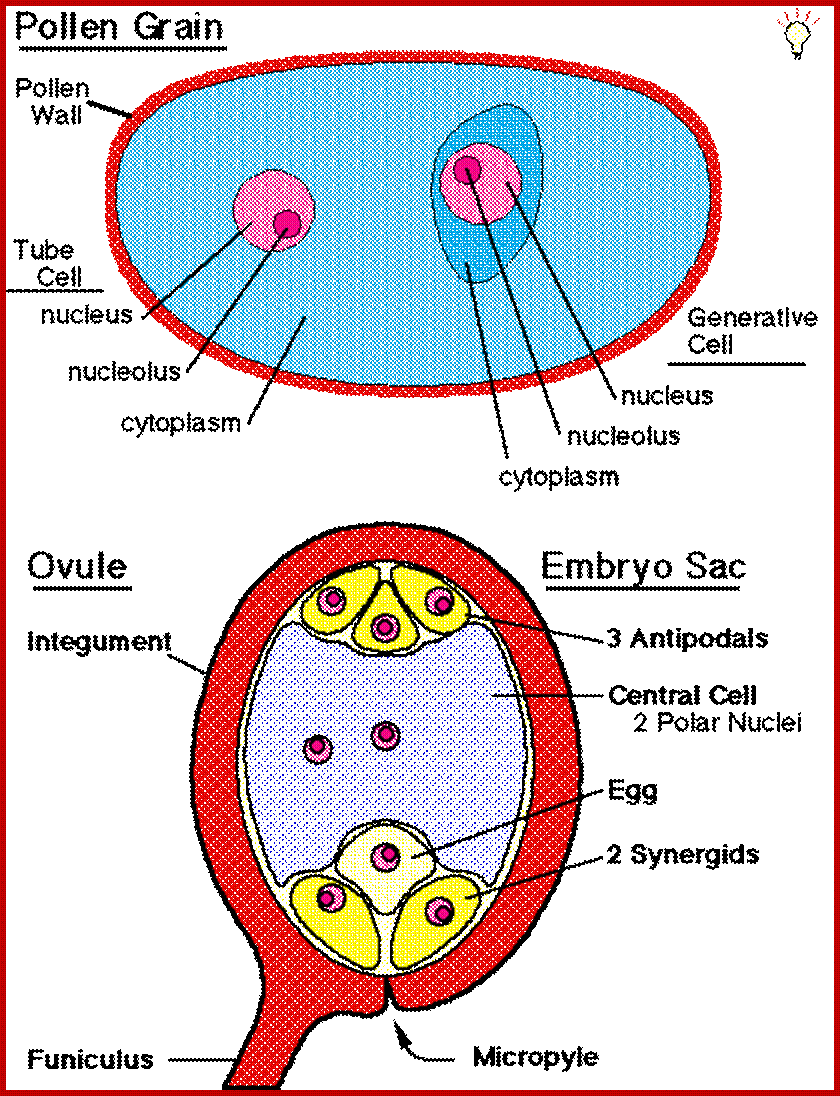
Flowers enclose reproductive Anthers and Ovules; anthers produce pollen gains (male reproductive component) and Ovules produce eggs female gametes; www.plantphys.info
Plants growing in different regions of the globe are exposed to different climatic conditions and different day length periods. In fact they are adapted to environs in such a way, they exhibit alternate vegetative and flowering cycles. That means that plants with their inherent genetic potentiality interact with environmental conditions; accordingly, they respond and behave. Humans, early modern humans Homo sapiens sapiens detected by fossil records (1868) of flower remains in 23,000 to 27000 old Cro-Magnon rock shelter site near a village Les Eyzies. Homo sapiens today, just about 45000yrs to 60000yrs old, copulated with Homo eructus, mostly in Asia and made them extinct; when they evolved and colonized sites of their own. These animals after many many centuries the above said species devised different methods to cultivate crop plants in different seasons of the year, so as to get the harvest at the right time of the year. Most of the mammals originated and developed were plant eating animals. They also domesticated many animals and plants (Agriculture) for their own use.

http://anthro.palomar.edu/
The common knowledge of the farmer has been extended and explained by plant physiologists; why and how the said plants behave in response to different environmental conditions. Plants have all the needed signal transduction pathways to respond to environmental signals. Such signal pathways has been worked out in Arabidopsis. Plants genomes and their physiology related to their molecular aspects are intelligently designed for the plants to prosper and survive to different and difficult climatic and environmental changes.

Developmental pathway of plants and its structures start from the zygote and end up in fully formed structures. Arabidopsis thaliana; This plant has been studied extensively at molecular and biochemical level. www. seattlecentral.edu

Some Parasite like bacteria turn a plant into flowerless Zombies; www.iflscience.com

Biological Process of growth and flowering fruiting; Arabidopsis; http://www.mun.ca/

Tunica Corpus the basic cell organization for the development of plant shoot system; L1 and L2 are epidermal and subepidermal layers respectively and the inner L3 is corpus https://en.wikipedia.org.

A schematic depiction of the organization of the SAM: A, Radial domains. Lateral organs are produced from cells recruited from peripheral zone, whereas cells from the rib zone contribute to the bulk of the stem. The central zone acts as a reservoir of stem cells that replenishes the cells of the peripheral and rib zones, which are lost during the formation of stem and lateral organs. At the same time, the central zone also maintains the pool of cells for itself. B. The clonally distinct layers of cells; The epidermal (L1) and sub epidermal (L2) layers maintain their distinctness by anticlinal cell division. The L1 and L2 layers are collectively referred to as the tunica. Cells interior to the L2 constitute the corpus (L3) in which cell divisions take place in various planes, resulting in growth in all directions. www.plantphysiol.org

View of the SAM; quiescent center; http://www1.biologie.uni-hamburg.de/

Beyond the Divide: Boundaries for Patterning and Stem Cell Regulation in Plants;;http://journal.frontiersin.org/

www.pcb.org

Cell division pattern like Periclinal; Parallel to the outer surface; anticlinal-parallel to lateral cell; http://www.mun.ca/biology

www.mun.ca

Shoot meristem central zone (CZ) , the peripheral zone(PZ); three layers L1, L2 and L33 are shown. Stem cells CLV3 and organizing center WUS homeobox gene interact in a negative feedback loop. The leaf primordial adaxial and abaxial cell fates are marked by expression of HD-ZIP III and KANADI family of genes; http://journal.frontiersin.org/
In dicot angiosperms the shoot meristem is organized into outermost layer L1 and its underlying cell layer called L2, both divide periclinally and give rise to the epidermal and subepidermal layers of cells. But the L3 cells divides in all directions giving rise to inner tissue. Clonal studies have determined that all post embryonically formed shoot cells ultimately are derived from about three stem cells in each layer (Stewart and Dermen, 1970). They are located in the outermost area of the central zone (CZ) that is defined by a lower cell division rate, compared to the peripheral zone (PZ) where lateral organ anlagen are initiated, and the underlying rib zone (RZ) that forms the pith tissue (Lyndon, 1998).
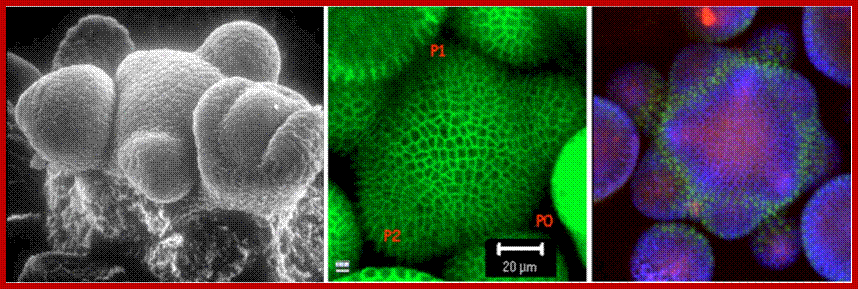
Arabidopsis shoot apical meristem. Left, scanning electron micrograph of the shoot apical meristem as it produces flowers on its flank (see Smyth et al. 1990). Center, a frame from a live-imaging movie in which plasma membranes have YFP inserted (Reddy et al., 2004). Right, shoot apical meristem of a triply transgenic plant showing REVOLUTA (red), PINFORMED 1 (blue) and KANADI (green, see Heisler et al., 2005).;http://plantlab.caltech.edu/

Schematic of genes that regulate stem cell activity in the Arabidopsis shoot apical meristem www.plantbio.berkely.edu
Molecular cloning of the genes showed that the central zone synthesizes a peptide ligand, CLAVATA3, which is secreted from the central zone cells, and activates a transmembrane receptor kinase, CLAVATA1, in rib meristem cells, eventually causing reduction in the size of the rib meristem (Fletcher et al. 1999). Computational morphodynamics (Elliot Meyerowitz) has been employed for live imaging of gene function and of protein levels and subcellular locations by real-time imaging. Plant scientists are currently analyzing several plant stem cell maintenance pathways. The Arabidopsis CLAVATA3 (CLV3) gene encodes a small-secreted polypeptide that is expressed in the shoot and floral stem cells and perceived by several receptor complexes at the surface of the underlying cells. Intercellular signaling through the CLV3 pathway restricts stem cell accumulation by limiting the expression of the WOX family transcription factor gene WUSCHEL (WUS), which in turn promotes stem cell fate and directly activates CLV3 transcription. This regulatory pathway functions as a negative feedback loop that maintains a functional balance between stem cell accumulation and organ formation throughout the plant life cycle.
In addition to the CLV3 pathway, plant scientists have identified the Arabidopsis ULTRAPETALA1 (ULT1) locus as an important negative regulator of shoot and floral stem cell activity. ULT1 encodes a SAND domain putative transcriptional regulator that restricts stem cell accumulation and operates as a critical timing component of a pathway that terminates stem cell fate during flower formation. Plant scientists have demonstrated that ULT1 acts as a trithorax Group (trxG) factor that regulates the chromatin conformation of large numbers of target gene loci. Present efforts are to further characterize the biochemical properties and downstream targets of ULT1 and the related ULT2 protein, and to identify additional components of the pathway.
Discovery of flowering response:
Though it is a common knowledge that different kinds of plants respond to different seasons of the year and produce flowers, it was left to G.Gassner & W.W. Garner to explain the phenomenon by their pioneering scientific studies. Gassner observed that winter variety of Petkus rye plants called Secale cereale, responded favorably to cold treatments. Almost at the same period of time, Garner and Allard demonstrated how plants produce flower in response to different lengths of the day and night in a 24 hours day cycle. The above two phenomenon are popularly called as Vernalization and Photoperiodism respectively. The above studies have led to the discovery of how plants rhythmically respond and behave to the day and night duration or to temperature fluctuation in different seasons of the year and they also observed rhythmical behavior of the plants which is referred to as ‘biological rhythm’ or circadian rhythm. And the operational time measuring system found within the plant structures, it is called ‘Biological Clock’.
Developmental Signaling;
The use of functional genomics to characterize members of a plant-specific family of CLV3-related signaling molecules called Clavata 3/ESR-related) CLE proteins (have 14 a.a motifs) and determine their roles in plant development. Intercellular signaling pathways convey cell fate information, regulate cell division and differentiation processes, and propagate and amplify specific signaling states. Yet members of only a few families of plant small signaling molecules have been studied and very little is known about how they coordinate growth and development. They have determined that most Arabidopsis tissues express multiple CLE genes in highly specific patterns, indicating that CLE-mediated signaling pathways are likely to play roles in many biological processes. This work has also demonstrated that, like CLV3, the CLE proteins function as secreted polypeptides that act in diverse intercellular signaling modules along with other WOX family members. We are currently studying the roles of several CLE polypeptides in Arabidopsis shoot apical meristem function and leaf formation. SAM consists of OC organizing center. In root tips one finds QC (quiescent center) equivalent to OC; Jennifer Fletcher
Expression of FT gene: Its regulation;
FLOWERING LOCUS T (FT) mol.wt 22kDa, similar to rice Hd3a (orthologous) is made in the companion cells of the leaves in response outside signals; and it is transported from the leaves to the apical meristem through the phloem sieve elements. FT is not species specific, by grafting it can induce flowering in plants across species. Recently, movement of FT from the companion cells to the sieve elements was shown to require the interaction between FT and a novel endoplasmic reticulum membrane protein called FT-INTERACTING PROTEIN 1 (FTIP1). TWIN SISTER OF FT (TSF) is a closely related protein and probably acts in a similar way to FT.
The proximal 5' region of the FT promoter is the binding site for many transcription factors which can repress or activate FT in response to external parameters. CDFs and TEMs are able to repress FT expression and are regulated by the circadian clock. CIB activators are able to induce FT under blue light enriched conditions. SVP is able to repress FT expression under low temperature, and PIF4 increases it under high temperature. This regulatory region however, is normally not accessible to transcription factors through the activity of LHP1, which is enriched in this region, and PRC2, which is able to trimethylate lysine 27 residues on histones in this region. Only the -5.3kb CCAAT box motif is free from the actions of LHP1-PRC2. Once NF-Y factors are able to bind to the upstream CCAAT box sites and recruit CO to the FT locus, CO activity is able to remove the LHP1 presence in the 5' proximal region, which enables the activity of other FT regulators. EFM and JMJ30 form a complex that regulates FT through demethylation of histones in the FT locus. MADS domain factors FLC, SVP, FLM, and MAFs, are able to bind to the first FT intron to repression transcription, both in response to low ambient temperature as well as prior to vernalization. AP2 repressors TOE1, 2, SMZ and SNZ are able to bind to the 3’ regulatory region near the FT 3' UTR to regulate FT expression.
FT activation occurs through two mechanisms of CO, the first being direct binding to CO-responsive elements (CORE) in the FT promoter and the second is recruitment of additional proteins that compose CO activator complex to assist transcriptional activation.

Transcriptional regulation of FT gene by CO proteins; http://www.ncbi.nlm.nih.gov/
At the shoot meristem, genetic data indicate that the FT and FLOWERING LOCUS D (FD) complex activates expression of flowering genes shown as a network in the meristem. In ft tsf double mutants and fd mutants, transcription of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) in response to long days is delayed in comparison to wild-type plants. SOC1, which encodes a MADS box transcription factor (56 to 60 a.a with DNA binding domain), is the earliest gene shown to be upregulated in response to long days at the meristem, and mutations in the gene cause late flowering. Strikingly, inactivation of other SOC1 and the related MADS box gene FRUITFULL (FUL); also known as AGL8 almost entirely suppresses the extreme early flowering caused by overexpression of FT from different heterologous promoters, suggesting that SOC1 and FUL are essential for the promotion of flowering by FT. When SOC1 is expressed in the meristem, it interacts with AGL24, another MADS box transcription factor, and promotes the activation of transcription of LEAFY (LFY), which is a meristem identity gene that is involved in the initiation of flower development. SOC, FUL, LFY and APETALA1 — another class of floral meristem identity genes — are also activated by SQUAMOSA BINDING PROTEIN LIKE (SPL) transcription factors, which are expressed in the meristem in response to FT and FD and were recently proposed to be direct targets of FD on the basis of chromatin immunoprecipitation experiments. Interestingly, SOC1 also binds to the SPL genes, suggesting that FD might act at different layers of the hierarchy to upregulate both SOC1 and SPL gene transcription (www.Nature.com). Interestingly scholars have found an important role for miRNA172 which targets MZ (Schlafmutze) a potent repressor of flowering.
Similarly, the FT–FD complex has been proposed directly to activate AP1, which is expressed in the cells that will give rise to the flower and confer floral identity on this primordium. However, recent detailed analysis of the AP1promoter questioned whether FD binds directly or not. Nevertheless, in rice, FD was also proposed to bind directly to the promoter of the AP1 homologue MADS15 via a similar element. The expression of both meristem identity genes AP1 and LFY is antagonized by TERMINAL FLOWER1 (TFL1), which is a protein that is related to FT, preventing their ectopic expression in the centre of the shoot meristem. In young floral primordia, AP1 and LFY repress TFL1 transcription. Recently, TFL1 was also shown to depend on FD to trigger the transcriptional repression of its targets, suggesting a pivotal role of FD, depending on whether it interacts with FT or TFL1 http://www.nature.com/.

http://vle.du.ac.in/
www.Nature.com
Genetic netwoerk of floral transition at the shoot apical meristem; FLOWERING LOCUS T (FT) is made in the companion cells of the leaves and is transported from the leaves to the meristem through the phloem sieve elements. Recently, movement of FT from the companion cells to the sieve elements was shown to require the interaction between FT and a novel endoplasmic reticulum membrane protein called FT-INTERACTING PROTEIN 1 (FTIP1)40. TWIN SISTER OF FT (TSF) is a closely related protein and probably acts in a similar way to FT. At the shoot meristem, genetic data indicate that the FT–FLOWERING LOCUS D (FD) complex activates expression of flowering genes shown as a network in the meristem. In ft tsf double mutants and fdmutants, transcription of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) in response to long days is delayed in comparison to wild-type plants34, 72. SOC1, which encodes a MADS box transcription factor, is the earliest gene shown to be upregulated in response to long days at the meristem, and mutations in the gene cause late flowering115, 116. Strikingly, inactivation of both SOC1 and the related MADS box gene FRUITFULL (FUL; also known as AGL8) almost entirely suppresses the extreme early flowering caused by overexpression ofFT from different heterologous promoters, suggesting that SOC1 and FUL are essential for the promotion of flowering by FT117, 118. When SOC1 is expressed in the meristem, it interacts with AGL24, another MADS box transcription factor, and promotes the activation of transcription of LEAFY (LFY), which is a meristem identity gene that is involved in the initiation of flower development119. SOC, FUL, LFY andAPETALA1 — another floral meristem identity gene — are also activated by SQUAMOSA BINDING PROTEIN LIKE (SPL) transcription factors, which are expressed in the meristem in response to FT and FD and were recently proposed to be direct targets of FD on the basis of chromatin immunoprecipitation experiments120, 121, 122. Interestingly, SOC1 also binds to the SPL genes, suggesting that FD might act at different layers of the hierarchy to upregulate both SOC1 and SPL gene transcription121. Similarly, the FT–FD complex has been proposed directly to activate AP1, which is expressed in the cells that will give rise to the flower and confer floral identity on this primordium45. However, recent detailed analysis of the AP1 promoter questioned whether FD binds directly123, Nevertheless, in rice, FD was also proposed to bind directly to the promoter of the AP1 homologue MADS15 via a similar element46. The expression of both meristem identity genes AP1and LFY is antagonized by TERMINAL FLOWER1 (TFL1), which is a protein that is related to FT, preventing their ectopic expression in the centre of the shoot meristem. In young floral primordia, AP1 and LFY repress TFL1 transcription124, 125. Recently, TFL1 was also shown to depend on FD to trigger the transcriptional repression of its targets126, suggesting a pivotal role of FD, depending on whether it interacts with FT or TFL1.www.nature.com

Identification of three classes of genes that control specification of floral organs in Arabidopsis
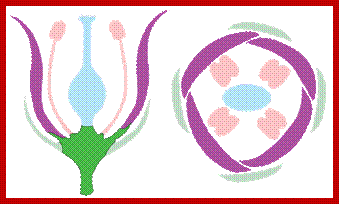
(a) Schematic diagram of the arrangement of wild-type floral organs, which are found in concentric circles called whorls. (b) Effect of loss-of-function mutations leading to transformations of one organ into another. Class A mutations affect organ identity in whorls 1 and 2: sepals (green) become carpels (blue) and petals (orange) become stamens (pink). Class B mutations cause transformation of whorls 2 and 3: petals become sepals and stamens become carpels. In class C mutations, whorls 3 and 4 are transformed: stamens become petals and carpels become sepals. [See D. Wiegel and E. M. Meyerowitz, 1994, Cell 78:203.]; http://www.ncbi.nlm.nih.gov/


Top left- to right-A,B, bottom left to right-C and D; http://www.theprogressplant.com/

http://www.theprogressplant.com/
A
mutants; If the ‘A’ gene is
removed, C fills it’s space in whorl 1 and 2 but without the influence of the
‘A’ gene the sepals and petals are transformed into more stamen and carpals.; B mutants; If we
remove ‘B’ we leave only ‘A’ and ‘C’; we get double sepals instead of petals,
and double carpals instead of stamen.; C
mutants; If we remove ‘C’, ‘A’ takes over its space and we get
a flower made entirely of sepals and petals (quite lovely in my opinion, but
unable to reproduce). ‘E’
Mutants; E-type mutants can be made by either removing all
three genes (A, B and C) or by removing the E-type genes that form the base for
the others to function. The plant is still getting the signal to make a flower,
but has none of the instructions to make the different organs. Its a bit like
trying to assemble a piece of Ikea furniture without the instruction-sheet or
allen key. The plant knows it’s time for flowers so gives its best shot with
what it’s got. A funky thing that looks like a flower, but is made entirely of
leaves..

The ABC model of floral organ development and the role of SCFUFO in regulating the B-type gene AP3. The diverse roles of ubiquitin and the 26S proteasome in the life of plants; James A. Sullivan, Ken Shirasu & Xing Wang Deng; In the 'ABC' model of floral organ development, organ identity is controlled by the expression of three classes of genes (A, B and C) in overlapping domains. In the first whorl, A-type genes are expressed alone and produce sepals. In the second whorl, the co-expression of A- and B-type genes results in the production of petals. In the third whorl, the co-expression of B- and C-type genes produces stamens, whereas the expression of C-type genes in the central whorl produces carpels. The SCFUFO has a key role in controlling the activation of the B-class genes, such as AP3 (but not PI), in the second and third whorls. However, so far, neither the ubiquitylation activity nor any substrate has been identified, although a role for the downstream regulator LFY has been indicated. AP3, APETALA3; LFY, LEAFY; PI, PISTILLATA; UFO, UNUSUAL FLORAL ORGANS. http://www.nature.com/nrg/

Expression patterns for three classes of floral organ – identity genes in wild-type Arabidopsis (a), loss-of-function mutants (b), and a transgenic that mis expresses class B genes (c) Colored bars represent the A, B, and C mRNAs in each whorl (W1, W2, W3, W4). The observed floral organ in each whorl is indicated below: sepal = se; petals = pe; stamens = st; and carpels = ca. See text for discussion. [See D. Wiegel and E. M. Meyerowitz, 1994, Cell 78:203; B. A. Krizek and E. M. Meyerowitz, 1996, Development 122:11.]; http://www.ncbi.nlm.nih.gov/

A landmark accomplishment in plant developmental biology is the ABC model of flower organ identity; http://www.nature.com/

Four floral organs sepals whorl1, petals whorl2, stamens whorl3 and carpels whorl4; ‘A’ class genes identify sepals; A and B identify petals, B and C class identify stamens, and C class identify carpels; APETALA1 (AP1 and AP2, B class genes are APETALA3 and PISTILLATA and C-class genes are AGAMOUS. Except AP2 all other homeotic genes code for MADS-domain proteins; the floral quartet model was proposed to explain the molecular mechanism of action underlying ABCDE protein function. Organ specific combinatorial quaternary MADs-domain protein complexes are proposed to control differentiation and outgrowth of distinct floral organs in four concentric rings/ whorls ( http://dev.biologists.org/).

Model for the action of MADS-domain protein complexes; In this model MADS-domain protein complexes forming regulatory proteins. The MADS green and blue form a quartet complexes and interact with two DNA binding sites (CArG boxes-Black-CC[A/T]6GG) in close proximity, resulting in DNA looping. The MADS domains recruit transcriptional co-factors (pink) which mediate transcriptional regulation and may influence target genes specificity as well as chromatin remodeling protein (brown), which relax chromatin at the target gene transcription start site allowing initiation of transcription.www.dev.biologists.org

How genes paint flowers and seeds; Mutant analyses have given insight into the various parameters that contribute to flower colour and pattern, which is so important for pollination. One important factor is the accumulation of orange, red and purple anthocyanin pigments in the cell vacuole—patterns arise by cell-specific expression of combinations of regulatory proteins. The overall colour perceived is also influenced by vacuolar pH, co-pigmentation and the shape of the petal cells. Although understanding of the biochemistry and genetics of anthocyanin and flavonol biosynthesis is well developed, this is not the case for pH and cell-shape control. www.cell.com
PHOTOPERIODISM (PP):
Earth, because of its rotation on its own axis and orbiting around the sun, exhibits a periodic day and night and seasonal changes. The duration of the day and night again shows variations because of the angle and distance between the earth and the sun at any given time of the year. This has an effect on temperature changes season wise. Thus plants and animals living on different parts of latitudes or longitudes are subjected to different periods of photo periods and different temperatures at different seasons of the year.
If we use three points or places on the globe, located at different positions as the reference point, to measure the day and night periods, it will be apparent how different are the day periods and temperatures of such places. Brazil in South America and Congo in Africa exhibit almost 12 hours of day and 12 hours of night in all the months of a year. But a city like Philadelphia located in the east coast of USA at latitude of 40 degree N, in the month of December; it experiences 9 hours of day and 15 hours of night. On the other hand, in the month of June, the day period is 14 ½ hours and night is 9 ½ hours long. Similarly, cities in Norway, during December, experience 6 hours of day and 18 hours of night, but in June, it enjoys 18 hours of day and 6 hours of night. Such day periods also accompany with changes in extreme temperatures. The above observations suggest that organisms living in these regions are subjected to seasonal variations of day and night and also to changes in seasonal temperature fluctuations.
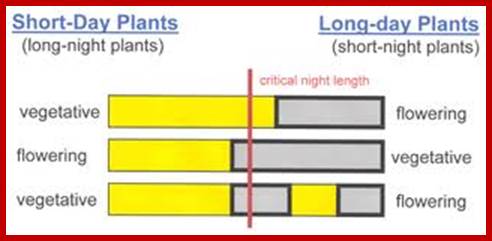
Garner and Allard, while working in the department of Agricultural Station, Beltsville, Maryland, USA, demonstrated remarkable relationship between the effect of the day period and flowering in a mutant tobacco plant called Maryland Mammoth. They observed that the mutant failed to produce flowers but grown tall, so they called it Maryland Mammoth. But the same plant started flowering when transferred to green house where it was subjected to short day and long night conditions. So the plant was called it a short day plant. Since then, a large number of plants have been subjected to various cycles of photoperiod i.e. treatment and according to their responses, plants have been classified into different groups. The flowering response in plants to photoperiodic treatment is now called photoperiodism. Light induced responses in photo morphogenesis are many and intricate; and this can be only represented in the form of network.
Classification:
Based on the responses to different photoperiods, most of the plants are grouped into 3 major classes, viz. Short day plants, long day plants and day neutral plants. However, detailed studies on each of these groups resulted in further classification of them into sub groups like long short day plants, short long day plants etc. Each of these groups has been further grouped into qualitative and quantitative varieties based on the specificity of the appropriate light periods.
Critical Day Period:
It is the duration of the photoperiod or the dark period that ultimately determines whether the plant has to go through vegetative growth or to produce flowers. Different plants require different periods of light or dark for 100% flowering. If that period falls short then plants do not produce 100% flowering. Such requirement of a minimum of photoperiod or dark period for effective flowering is called critical day period. The length of light and dark period for different long day and short day plants varies. For example Xanthium requires a critical length of 15 ½ hours of dark period for its effective flowering. If the dark period is less than 15 ½ hours plants do not induce any flowering, but longer dark periods d not inhibit flowering. On the other hand, the long day plant, ex. Hyoscyamus niger requires a critical 11 hours of exposure to light. Anything less than that, plants fail to produce flowers. If the length of the day period for this plant is more than 11 hours, it does not affect the flowering. Similarly, different plants have different critical day periods and the correct photoperiod has to be determined individually by subjecting them to photoperiodic treatment. Plants have built in genetic processes to produce flowers.

www.photoperiodism.brainstars.org
Ripeness to flowering and site of perception:
Not all photoperiodic plants respond to the light treatment until and unless the plant has grown to certain vegetative maturity. For example, Wulfia requires at least one leaf; Xanthium responds well if it has few partially mature leaves. In Zea mays, at least there should be 5-6 leaves to respond for photo periodic treatment.

www.anglerz.com

This model is based on microscopic analysis of constituent cells. It says that the shoot apical meristem is made up of two groups of cells. The tunica, a group of cells that form one or two stratified layers, undergoes anticlinal divisions only and gives rise to the epidermis.
Partly enclosed by the tunica is the corpus, a group of loosely arranged cells that divide in various planes and give rise to the vascular and ground tissues. The tunica maintains its individuality by surface growth, whereas the corpus adds bulk by increase in volume.
http://lifeofplant.blogspot.in/

https://learning.uonbi.ac.ke
Stem apex is the site for development of flower; a dramatic change in the structural and functional features of the SAM takes place. Floral organs are nothing but modified shoot system. The floral structures are same as the vegetative structure but modified; sepals, petals, stamens and carpels are all derived from leaf primordial. The most intriguing question that has puzzled scientists for such a long time is that, how the genes hitherto remain silent in these primordial, start expressing, differentiate and develop into floral structures. The molecular aspect of gene expression that changes leaf primordial into floral organs is revealed but not completely.
 The genetic basis of flowering responses to seasonal cues;
The genetic basis of flowering responses to seasonal cues;
Plants respond to the changing seasons to initiate developmental programmes precisely at particular times of year. Flowering is the best characterized of these seasonal responses, and in temperate climates it often occurs in spring. Genetic approaches in Arabidopsis thaliana have shown how the underlying responses to changes in day length (photoperiod) or winter temperature (vernalization) are conferred and how these converge to create a robust seasonal response. Recent advances in plant genome analysis have demonstrated the diversity in these regulatory systems in many plant species, including several crops and perennials, such as poplar trees. Here, we report progress in defining the diverse genetic mechanisms that enable plants to recognize winter, spring and autumn to initiate flower development.;www.nature.com

Flowering is controlled by a variety of interrelated mechanisms. In many plants, the environment controls the production of a floral stimulus, which moves from the leaves to the shoot apex. Apices can become committed to the continuous production of flowers after the receipt of sufficient amounts of floral stimulus. However, in some plants, the commitment to continued flower production is evidently caused by a plant’s commitment to perpetually produce floral stimulus in the leaves. Ultimately, the induction of flowering leads to the specification of flowers at the shoot apex. In Arabidopsis, floral specification and inflorescence patterning are regulated largely by the interactions between the genes TERMINAL FLOWER, LEAFY andAPETALA1/CAULIFLOWER.

Looking at the start of the plant body from the zygote a fertilized egg, in course of time, it divides and redivides and differentiates and develops tissues and organs. Developmental process in plants, at molecular level is not same as in animal systems, but use different set of gene expression and different pathways. However homeobox genes are involved in both.
The figure below shows an inflorescence shoot apical meristem (SAM) and two adjacent floral meristems (FM) of Arabidopsis thaliana. On the left is the original laser scanning confocal microscope optical section of tissue stained with propidium iodide to show the nuclei. The center image was colored to show radial zonation within the SAM. The central zone (CZ) is shown in red, the peripheral zone (PZ) in green, and the rib meristem (RM) in blue. The image on the right was colored to show clonally-related layers. The epidermal L1 layer is shown in blue, the sub epidermal L2 layer is shown in pink, and the L3 layer, or corpus is shown in gold. The L1 and L2 together are called the tunica, www. http://biology.kenyon.edu/


Homeotic genes control organ identity
Shoot meristem converts to inflorescence
meristem which
can form one or more floral meristems.
The floral organ primordia arise from floral
meristem by cell differentiation and enlargement.
4 concentric whorls reflect the order within the
floral meristem. Sepal (whorl 1) from the outer ring. Petals (whorl, 2) from the next ring. Stamens (whorl 3; male reproduction) from the
inner ring. Carpels (whorl 4; female reproduction) from
the centre. In Arabidopsis there are 15 separate primordia (4
sepals, 4 petals, 6 stamens and 1 pistil [with 2 carpels]).http://www.mun.ca/
Look at the Stem Apical Meristem (SAM), it has undifferentiated central dome of progenitor cells (pleuripotent in nature) covered by a layer of epidermal cells. Signals have to come from different modes and methods to convert such potent cells to go through developmental programmes. It is possible that one such cell is enough for the development, just like stem cells in animal systems. Signals are varied such as sunlight, sunlight duration, temperature, and organic chemicals, hormones such as Gibberellins and food sources as internal signals. Cellular transduction, in response different signals, is more complex, for the signals arrive as environmental factors; light (Photoperiodic pathway) temperature (Vernalization pathway) or inbuilt factor (autonomous pathway). Most of the environmental signals impinge on leaves and resultant downstream products have to be translocated to the SAM; this can be long distance for the site of perception and the site of response are separated in time and space (Einstein).

Structure of the root (A) and shoot (B) stem cell niches. Both are composed of an organizing center (OC; referred to as the quiescent center in the root), which maintains stem cell identity in a neighboring population of cells. (C) The regulatory signaling network controlling the identity of the organizing center and stem cells in the shoot meristem. (D) The immediate effect of suppressing CLV3 expression is the expansion of stem cell identity beyond the central zone of the shoot meristem into the surrounding peripheral zone. http://www.sciencedirect.com/

Maintenance of stem cells in shoot apical meristem; The SAM is organized in three functional zones [central zone (CZ), peripheral zone (PZ), and rib zone (RZ)] and three layers where the antagonistic relation between WUS and CLV is essential to preserve cells in the meristem. WUS activates CLV3, which further binds with CLV1/2 and in turn inhibits expression of WUS. Cytokinin positively controls WUS expression where ARRs are negative regulators of cytokinin and are inhibited by WUS. The L1 specific miR394 negatively affects the LCR protein, which interferes in WUS/CLV based stem cell maintenance (pointed and T shaped arrows indicate positive and negative regulation, respectively). http://journal.frontiersin.org/

Developing shoot apical meristem (SAM) in barley; Different developmental phases can be distinguished based on morphological changes of the SAM. www.mpipz.mpg.de
Plants without leaves do not respond to any photoperiodic treatment, which suggests that the vegetative buds alone or perse are incapable of perceiving the stimulus? Similarly, plants with only old and mature leaves not only fail to respond but also they inhibit or nullify the photoperiod effect. However, partially mature leaf or leaves that are just unfolding, are highly sensitive. A remarkable feature is that even one such sensitive leaf is enough to respond to proper photoperiodic induction. It means whatever reactions or a product produced in one leaf is enough to induce flowering apical meristem wherever they are present. Let us start with light mediated induction.
Light and Duration mediated:
Photo inductive Cycle:
A plant, if subjected to certain length of day period and night period in 24-36 hours duration, then it is called one photoperiodic cycle. Such periodic cycle responsible for inducing flower formation, is referred to as ‘photo inductive cycle’. The required number of photo inductive cycles varies from species to species; this is because of inherent genetic makeup of the said plant. For example, Cockleburr needs just one photo inductive cycle but plants like Plantago lanceolata or Salvia accidentales require at least 17-25 cycles for 100% flowering. If the provided inductive cycles are less than what is required, the number of flowers produced in such plants will be less than 100% (a quantitative effect). This suggests that during the inductive cycles some flower inducing material gets accumulated and if such a substance produced reaches a threshold value the flower production is maximal.
It is important to know that the photoperiodic response is ‘all or none’ phenomenon. Once the provided stimulus produces a proper impact on the genome, the flowering is initiated and once it is initiated it cannot be reverted to vegetative condition under normal circumstances.
Importance of Dark Period:
Systematic studies, using different lengths of light and dark period, it is noticed that the dark period is important for short day plants for flowering. Shortening of photoperiod has profound influence on the quantitative yield of flowers. The importance of photoperiod, i.e. Day period on short day plants is very interesting. For example Xanthium requires 15 ½ of continuous dark period and 8 ½ hours of day period for 100% flowering. If such a plant that is maintained with 15 ½ long dark period and the day period is shorted by 2 or 3 hours, the total number of flowers produced will be significantly lower than the plant that is exposed to 8 ½ hours of day period. This suggests that the photoperiod affects flowering ability in quantitative terms, it means that proper dark period is essential for flowering but one cannot expect the plant to grow and produce flowers in continuous dark. This is because light is required for photosynthesis for it provides energy rich and components for the development of floral primordia. If the photosynthate supply is not adequate the total growth of the floral axis is affected. In fact, in one of the experiments, a short day plant which is kept in continuous dark conditions is induced to produce flowers by providing sugar solution to the leaves. Similarly if the CO2 supply is cut off during photosynthetic period, the short day plants in spite of receiving proper dark and light periods, they do not produce flowers to their maximum ability. The above observations suggest that photosynthate provides the necessary raw materials for floral primordia for full expression.

hoopermuseum; www.earthsci.carleton.ca
http://biology4isc.weebly.com/2-

The circadian clock occupies a central position in the regulatory network of plants. The traditional view has been of a linear pathway in which environmental stimuli, such as light and temperature, entrain the clock, which then regulate growth and reproduction; Kathleen Greenham & C. Robertson McClung; www.nature.com
Another important factor that affects the floral induction is the intensity of light. If the light provided is of low intensity i.e., less than 100 ft candles, flower production is totally inhibited, though the meristems are organized into floral primordia. But if the intensity of light is increased above a critical level, the number of flowers produced also increases up to certain level. This is because the light intensity affects the total yield of photosynthate, so flower production is also affected by the said factor.

http://biology4isc.weebly.com/
Action Spectrum of Light:
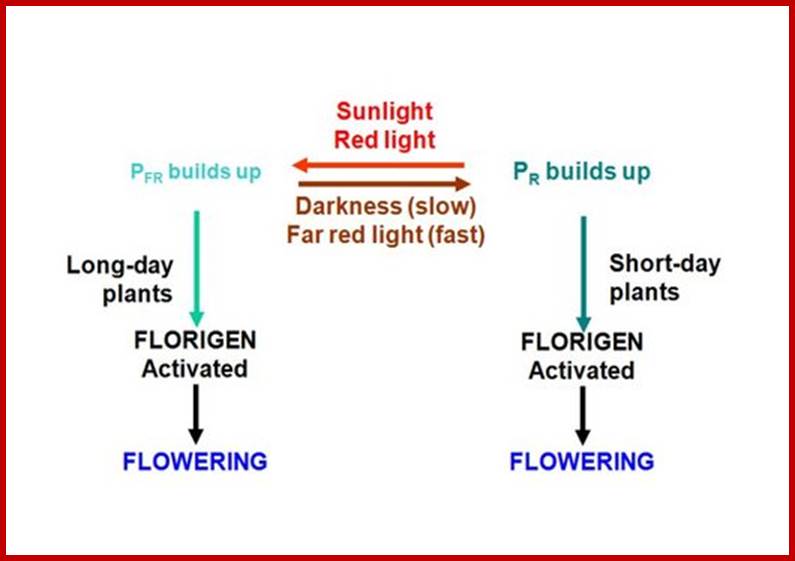
Finer analysis of the active part of white light that is effective is photoperiodic inductions reveals that the red light at 660 mm and far-red light at 730 mm are the most effective wavelengths in inducing or inhibiting the flower initiation. It has been established that continuous far-red irradiation inhibits flowering in long day plants, on the contrary, continuous red light treatment blocks flowering in short day plants.
The dramatic effects of red light and far red light can be demonstrated on a short day plant like xanthium. It is known that a short break in continuous dark period with white light in a short day plant brings about the total inhibition of flowering. If the short break is due to red light, the inhibition is 100%, but if the break is due to far red light, flowering is not inhibited. Interestingly, if the red light and far red treatment is repeated alternately but ending in Far Red as short breaks results in the reduction of total number of flowers produced. If the number of cycles is extended, the flowering will be totally inhibited though the last light treatment is Far-Red light. This is possibly due to the breakdown or exhaustion of some intrinsic factors generated during short day treatments.
Phytochrome as the Photoreceptor:
The effectiveness of red light and far red light inducing or inhibiting the induction of flowers strongly suggests the presence of some substance or substances that could absorb light in the said wavelengths. By absorbing light at a particular wavelength, the said substances probably undergo excitation of chromophore and the protein bound undergoes conformational changes. This protein complex induces signal transducing activities leading to flower induction. Search for such a compound in plants resulted in the discovery of blue / yellow colored pigments called Phytochromes.


The central hypothesis of phytochrome action, proposed almost 50 years ago from pioneering investigations by S. Hendricks, H. Borthwick and his colleagues, is that the photoreceptors exist in two, photoconvertible forms, Pr and Pfr. Pr is biologically inactive and upon absorption of red photons is converted to Pfr, the active form. Pfr is converted back to Pr by far-red photons. Biological action stems from Pfr.


Proposed structure for the Phytochrome-chromophore, its linkage to the
protein and possible phototransformation mechanism (after Rüdiger, 1972);
The 'blue' form is thought to correspond to P r and the 'green-yellow' form to Pfr.The
chromophore is an open-chain linear tetrapyrrole—known as phytochromobilin—and
is closely related to phycocyanobilin, the chromophore of the abundant algal
pigment C-phycocyanin.; Linear Phytochrome pigment; www. publishing.cdlib.org


Phytochrome is a linear tetrapyrrole is attached to a phytochrome protein; www.mobot.org and http://www.mobot.org/
![]()
The photo conversions involve a number of intermediate forms in both directions, and the establishment of equilibrium between Pr and Pfr takes several minutes even at daylight irradiance levels. The absorption spectra of the phytochromes peak at about 665 nm and 730 nm. The absorption bands overlap, so radiation below about 700 nm activates photo conversion of Pr and Pfr. Thus, in daylight for example, a photo equilibrium of about 60% Pfr/P (where P = total phytochrome) is established—in canopy shade or crowded communities, the photo equilibrium can be as low as Pfr/P = 0.1. This is the basis of the shade-avoidance syndrome.
The chromophore is an open-chain linear tetrapyrrole—known as phytochromobilin—and is closely related to phycocyanobilin, the chromophore of the abundant algal pigment C-phycocyanin. In higher plant phytochromes, the chromophore is covalently attached to the protein through a thio-ether link at a cysteine positioned at amino-acid residue 374 (numbering for phyA). Assembly of apoproteins and chromophore occurs spontaneously, presumably involving inherent chromophore lyase activity in the phytochrome apoproteins. This property has allowed the construction of recombinant phytochrome adducts with either phytochromobilin or phycocyanobilin—both are spectrally photoreversible and active when transgenically expressed in planta. Harry Smith
This complex when get excited with the absorption of light at red wave length, its proteins get activated and acts as an aspartate kinase and it autophosphorylates itself.
When phytochromes were discovered there was great excitement among plant physiologists. This led to intensive research work on various aspects of the structure and functions of phytochromes in plant morphogenesis. Now scientists have extracted and identified five phytochromes such as Phy A, B, C, D and Phy-E. Yet in recent years other pigment-protein complexes have been discovered, called Phototropins and Cryptochromes.
Distribution:
In recent years, the techniques for extraction and quantification of these subtle pigments have been standardized. The pigments have been found to be located in all possible organs of the plant body. At the cellular level, it is mainly associated with plasma membrane, cytoplasm and the membranes of plastids. The presence of this pigment in plastid membranes is significant, because plastids are also known to perform many other photo biological processes.
Chemical composition and structure:
The pure form of phytochrome appears blue / yellow colored pigment in solution form. Phytochrome, in fact, is made up of two moieties; one is a protein and the other is a chromophoric component. The phytochrome-associated protein has been isolated from maize seedlings and other sources. The mol.wt of the protein is 123-125 KD and it exists in tetrameric form. But chromophore part is made up of linear chain of four pyrole rings. The protein subunits are firmly bound to the A-pyrole ring via S-bond of the chromophore unit (chromophorobilin). With the absorption of light at 660 mm or 730 mm, the double bonds found within the chromophore get disturbed and shifted. These changes inturn bring about conformational changes in 3-D structure of the pigment and also in protein chains either in to trans form (activated) or cis form (inactive form). Probably, the above said changes due to absorption of light transform them into excited form of molecules and they inturn elicit certain physiological functions in the cells.

http://staff.washington.edu/


The two leaf like structures are dimeric phytochrome apoproteins.; www. www.scribd.com; www. seattlecentral.edu

Structural features of the phytochrome B protein, (there are 6 pytochrome molecules); indicating the positions of the tetrapyrrole chromophore attachment site, the ‘core’ region in the carboxy-terminal half of the protein, the two PAS-like domains and the histidine kinase-like domain (HKLD). The carboxy-terminal portion of the protein was fused to the Gal4 DNA-binding domain and used as ‘bait’ in a yeast two-hybrid screen that led to identification of PIF3 as a likely physiological partner; www.cell.com

plantphys.info; https://smartsite.ucdavis.edu

The Pr form abosorbing Red light changes into Pfr form and causes changes. The Pfr form of pigment-proteins also undergo conversion by their own slowly, otherwise absorption of far-red light changes them to Pr immediately. www.blog.gurukpo.com; https://smartsite.ucdavis.edu
Mol.Wt of Phototropin- 120kDa; http://biobook.nerinxhs.org/

http://pubs.rsc.org/
Phototropins are one of several classes of photoreceptors used by plants and algae to respond to light. These proteins contain flavin-binding LOV (Light-Oxygen-Voltage) domains that form covalent cysteine-flavin adducts upon exposure to blue light, leading to the enhancement of phototropin kinase activity. Several lines of evidence suggest that adduct formation in the phototropin LOV2 domains leads to the dissociation of an alpha helix (Jα) from these domains as part of the light-induced activation process. However, crystal structures of LOV domains both in the presence and absence of the Jα helix show very few differences between dark and illuminated states, and thus the precise mechanism through which adduct formation triggers helical dissociation remains poorly understood. Using Avena sativa phototropin 1 LOV2 as a model system, we have studied the interactions of the LOV domain core with the Jα helix through a series of equilibrium molecular dynamics simulations. Here we show that conformational transitions of a conserved glutamine residue in the flavin binding pocket are coupled to altered dynamics of the Jα helix both through a shift in dynamics of the main β-sheet of the LOV domain core and through a secondary pathway involving the N-terminal A′α helix.

LOV protein; http://www.biomedcentral.com/

Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction; Adiantum, Phy3 domain and LOV2 structures; ; Phy3 domain showing the N-termini of Phytochrome domain bound to Phototropin,; http://www.pnas.org/

https://commons.wikimedia.org

Phytochrome A and Phytochrome B Have Overlapping but Distinct Functions in Arabidopsis Development; Jason W. Reed et al
Dual forms of phytochromes:
In the earlier days of its discovery, people suspected the presence of two kinds of phytochrome pigments because they showed different absorption spectrum at 660 nm and 730 nm. But Norris and others, using dual wavelength spectrophotometer, demonstrated that the same phytochrome pigment exists in two alternate forms. They are called red light absorbing pigment and far-red light absorbing pigment. The Pr form after absorbing red light gets transformed into Pfr form which by absorbing Far-red light gets converted back to PR form. The Pfr form naturally undergoes decay back to Pr form. Thus phytochrome exhibit dual forms. The concentration of each form is dependent upon quality of the light source, duration of exposure and the physiological state of the cell.
The Pfr form of the pigment formed due to the absorption of red light is not very stable. It decays back to Pr form or it is degraded by some enzymes in dark condition, but this process is slow. On the other hand, if the PFR form absorbs far red light, it gets converted to PR quickly. But the decay of PfR pigment to PR form on its own takes place in dark or it is converted by certain enzymes and it is temperature dependent. In the presence of oxygen, the pigment undergoes irreversible destruction. Inspite of their labileness and sensitivity, they remain quite stable at pH 6 and pH 8. Furthermore, the stability of these forms of pigments is controlled by the firm binding of protein moiety to the chromophore part of the pigment.
Activated phytochrome undergoes autophosphorylation at serine/threonine residue, which then binds to its receptor protein and moves into the nucleus, where it associates with transcriptional factors and activate their target genes whose products inturn activate other genes and promote flowering.

Signal transduction; www.Science.com
LREs Light responsive elements, MYB
= TGGTTAG, G box= TGACACGTGGCA, GT1 = TGGTGGTTAATATG GATA motif= AAGATAAGATT

Absorption of a red photon by the inactive Pr form of a phytochrome causes a conformational change in the dimeric photoreceptor molecule. In the Pfr form, the phytochrome translocates to the nucleus where it binds to a putative reaction partner, PIF3, which is constitutively found in the nucleus and has the characteristics of a basic helix-loop-helix transcription factor. The Pfr-PIF3 complex initiates gene regulation, either directly or through unknown intermediates. Reversion of Pfr to Pr by far-red light results in rapid dissociation of PIF3, interrupting signal transduction. In the Pr form, the phytochrome slowly relocates to the cytoplasm. Here, in either the Pr or Pfr form, it can bind to a kinase substrate (PKS1), which may be involved in retention of the phytochrome in the cytoplasm or in its release for translocation. Phosphorylation of PKS1 is enhanced with Pfr, and may be the prelude to cytoplasmic action. So, several steps are susceptible to regulation by absorption of light by the photoreceptor: phosphorylation, nuclear translocation, association with PIF3 and transfer of signal transduction to PIF3.
;Main steps in Phytochrome action-Harry Smith; www.nature.com
Phytochrome in response to light undergoes conformational change and the Pfr moves into the nucleus where it binds to PIF3 that is already bound to the G-box (Light responsive element) of the responsive genes and activate the gene expression. Light also triggers phytochromes mediated G-trimeric membrane protein activation that leads to the production of cGMP and the release of Ca2+ ions. One of the early genes expressed in response to Phytochrome is activation of MYB (a family of TFs). The protein contains three domains, one N-terminal DNA binding domain, second-middle domain transcriptional activation domain and the C-terminal in transcriptional repression. The MYB in turn activates Circadian Clock Associated CCA1 gene, LHY (late elongated hypocotyl) gene and HY5 (Long Hypocotyl Elongation) gene (basic leucine zipper protein). In plants, the MYB (Myeloblastosis) family has selectively expanded, particularly through the large family of R2R3-MYB. Members of this family function in a variety of plant-specific processes, as evidenced by their extensive functional characterization in Arabidopsis (Arabidopsis thaliana). MYB proteins are key factors in regulatory networks controlling development, metabolism and responses to biotic and abiotic stresses. The elucidation of MYB protein function and regulation that is possible in Arabidopsis will provide the foundation for predicting the contributions of MYB proteins to the biology of plants in general. http://www.ncbi.nlm.nih.gov/
www.users.rcn.com

a |
Postulated direct targeting of light signals through phy molecules to a
promoter-bound basic helix–loop–helix factor (PIF3) that simultaneously
regulates both photomorphogenic and clock genes through a short, branched
transcriptional cascade. phyB translocates to the nucleus following
light-induced conversion of the Pr (PrB) to the Pfr (PfrB) form where it binds
to G-box-bound PIF3 and induces the expression of the primary target genes CCA1 and LHY. The encoded MYB-related transcription factors bind
in turn to their cognate binding sites (here, CCA1-binding site, CBS), where
they either induce expression of genes such as CAB or repress expression ofTOC1.
TOC1 is in turn a positive regulator of CCA1, creating a feedback loop that is postulated to
constitute the circadian clock (labelled![]() ). CAB, CHLOROPHYLL A/B BINDING
PROTEIN; PIC, PREINITIATION COMPLEX; PIF3, PHYTOCHROME-INTERACTING FACTOR 3;
TOC1, TIMING OF CAB EXPRESSION 1 PROTEIN. b |
Nucleotide sequences identified as a CCA1-binding site in the CAB gene promoter64, 66, and as a conserved motif (evening
element) and CCA1/LHY-binding site in a large group of clock-controlled genes68, 69. The pair of motifs in each case is a
closely related repeat that is separated by a spacer of eight nucleotides. www.nature.com
). CAB, CHLOROPHYLL A/B BINDING
PROTEIN; PIC, PREINITIATION COMPLEX; PIF3, PHYTOCHROME-INTERACTING FACTOR 3;
TOC1, TIMING OF CAB EXPRESSION 1 PROTEIN. b |
Nucleotide sequences identified as a CCA1-binding site in the CAB gene promoter64, 66, and as a conserved motif (evening
element) and CCA1/LHY-binding site in a large group of clock-controlled genes68, 69. The pair of motifs in each case is a
closely related repeat that is separated by a spacer of eight nucleotides. www.nature.com

PHY-Pfr phosphorylated form, not only moves into the nucleus but also interact with membrane trimeric alpha/beta/gamma G protein, that activates cGMP and leads to the release of Ca^2+. PHY binds to LHY/CCA1 gene LRE bound PIF3 and induce the expression of LHY and CCA1; they in turn activate TOC1 (Timing of Cab Expression 1) gene. The product of TOC1 in turn acts on LHY/CAA1;www.pcb.org

A model of phytochrome signal transduction. Activated phytochrome (Pfr) is proposed to regulate transcription through several parallel pathways. A rapid response involves Pfr translocation to the nucleus, where it binds transcription factors of the bHLH family (in particular PIF3). Key regulatory transcription factors (RTFs) that are responsible for inducing a range of light‐regulated genes are subsequently activated. In a second nuclear‐localized pathway, phytochromes are proposed to bind response regulators (RR), which stabilize them in the activated form and can induce light‐regulated gene expression by inhibiting COP1‐, COP10‐ and CSN‐dependent proteolysis of the HY5 transcription factor and by binding to activated cryptochromes (cry). In all cases, regulation of the genes responsible for photo morphogenesis is predicted to require chromatin remodeling mediated by the DET1/DDB1 nucleosome‐binding complex. In the cytoplasm, phytochrome may activate gene expression through G‐proteins (G), calcium and cGMP‐dependent pathways, which are regulated by SUB1. In addition, phytochromes may be sequestered away from the signalling‐competent pool by PKS1. Elements involved in signalling from specific photoreceptors or controlling specific responses have not been included. Involvement of Pfr-Phy-A and other in chromatin remodeling is speculated but not discerned. www. embor.embopress.org
Functions:
Probably phytochrome is the only pigment that is known to have multifarious activities and elicit diverse responses. Among the 35 to 45 functions attributed to this pigment-protein complex, bud dormancy, seed dormancy and flower induction are the well known phenotypic effects. At the metabolic level, the phytochrome is known to act upon cell respiration, permeability, transcription, translation and enzyme activity. Some of these activities have been very well demonstrated in different plant systems. In this text the discussion is restricted to its role in flower inductions and dormancy.
Role of phytochromes in flower induction:
Phytochromes being omnipresent in the plant body, they are always subjected to both red and far red radiations in the day conditions. Accumulation of pR forms and pFR forms of phytochrome in sufficient amounts in plants is critical and important. The effective concentration of any of these forms over a threshold values in the perceptive organs like leaves is absolutely essential to bring about certain biochemical functions which may ultimately lead to the induction of flowers.
In long day plants, the Pr form of the pigment by absorbing red light throughout the day transforms the substance to Pfr form and it accumulates in greater amounts. Such Pfr pigments, when preset in higher concentration above the threshold value, activate the cell machinery and ultimately induces flowering. However, recent studies indicate that the PfR form alone is not active, but it also requires another substance called X whose properties are not well characterized. The PER-X complex is believed to be highly effective in inducing flower formation in long day plants. The X factor is known to be Phytochrome interacting factor (PIF3).
PR – > PfR –> PRR.X – Induction of flowering.

Light activated Phy-A binds to the receptor FHY1-FHL (FHY stands for elongated hypocotyls or Long Hypocotyls) that enters the nucleus where it activates light response genes including flowering genes. Accumulation PhyA represses the production of FHY3-FAR1 for they bind to gene loci of the same. With the dissociation of Phy-A from FHY1-FHL the genes for FHY3 and PAR1 get activated to produce FHY1-FHL that is found in cytosol, which are used for the binding of Activated Phy-A. www.science20.com

Note that adult
tissue can be cultured and callus can undergo redifferentiation.
Transgenic plants can be generated in culture; http://www.mun.ca/biology
On the contrary in short day plants, because of long dark periods, whatever PfR pigments formed in the day conditions are subjected to decay back to PR form. However, higher levels of PR pigments, they are effective in inducing flower formation in short day plants. Conversely, higher amounts of PR forms inhibit flower initiation in long day plants and PfR forms prevent initiation of flowers in short day plants. So the kind of pigment or the form of pigment that has a promotive effect on one kind of plant acts as an inhibitor to the other kind. The dual form of pigments performing dual role is really intriguingly fascinating.
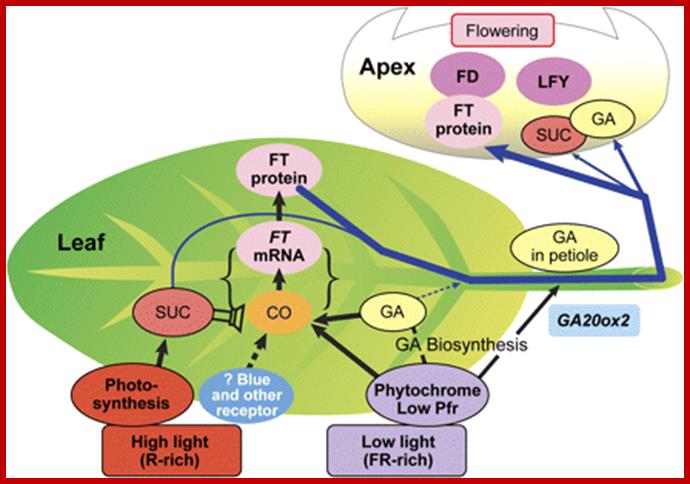
The PHY–PIF3 complex binds to LHY/CCA (long hypocotyls and Chlorophyll a/b binding proteins) region of the said genes and activate its transcription. The CCA has its own target genes such as TOC1 which in turn interacts with PHY-PIF3 dimer complex. The ultimate effect is on the synthesis of ‘flowering locus’ T (FT) in conjunction with constans (CO). Is this FT a protein or mRNA? This FT moves into phloem vessels that translocates to the base of SAM; www.pcb.org
This actually explains why gibberellins are effective on long day plants but not so in short day plants. Extrapolating this view it is possible to visualize that there are two or more different genetically regulated regulatory factors acting at two regulatory sites. Added to this, plants requiring vernalization can be short cut the flowering by GA treatment. But one thing is certain that one of the factors is GA and other factor, i.e. Anthesins may be anything, possibly it may be a highly labile protein or it may be one of the mRNPs, RNPs or a protein or signal factor or a kind of ligand that can induce signal transduction just binding to the receptors on plasma membranes or cytoplasmic receptor found in of the receiver tissues. The most enigmatic situation was GA actually synthesized in the apex of the future stem tip or inflorescence meristems, precursors for GA synthesis are found in proplastids and fully developed plastids. The photoperiodic stimulation takes place in young leaves; if so translocation of the phytochrome induced signal has to be transported a long distance to the non determinant stem meristem the future floral meristem. What is the structural element that can transport such substances? Is it Xylem or phloem? Xylem elements participation can be ruled out for the simple reason that the xylem elements transport is mostly from root to all other regions. But phloem transportation takes place in both directions; starting form vienlets to veins and to the midrib of the leaf and from there the transportation is bidirectional. Whether the bidirectional movement takes place in the same sieve tube-companion dimers or separate sieve tube elements is little ambiguous.
In addition to photoperiodic and GA pathway, plants use autonomous pathway and vernalization pathways also, where GA pathway provides a kind of interlink between photoperiodic and vernalization pathways.
Concept of Florigin:
The PR and PfR forms of pigments are the products of photoperiodic stimulus and they in turn are responsible for inducing flower formation. So the respective pigments elicit certain responses in the plants which probably produce some kind of a substance (s) which is/are responsible for transforming vegetative shoot into reproductive shoot. Production of such substances by plants has been suspected by many plant physiologists long ago. But so far, no one has succeeded in isolating or identifying such compounds. In spite of it, the presence of substance (s) responsible for flower induction has been proved by different methods and by different investigators.
The grafting experiments conclusively prove the presence of flowering substance(s). If a short day plant, kept under proper photo inductive conditions, is grafted to another plant or plants (by serial grafting) which are maintained under non photo inductive conditions, flowering is induced in all plants. Whether the plants receiving the graft are short day plants or long day plants, it does not make any difference. This experiment clearly indicates the production and existence of a flower inducing substance in a photo induced plant made product in response to stimulus, which is capable of diffusing through the graft to the receiver plant. What is this substance is it a chemical signal such as cAMPs like or is it an mRNA or a protein. Any substance that is induced and produced in leaf cells has to be transported long distances and should be stable. It is possible that the substance produced should cross through the cell wall of mesophyll cells into sieve tube cells, and then it has to be transported to stem apex meristems (SAMs). But now it is known it is synthesized in companion cells. There again it has to cross cell wall barriers to reach a whole mass of cells. So this substance should be a small molecule that can be easily transportable and easily induce signal transduction that is capable of spreading.
Chailkhyan, a noted Russian botanist named such flower inducing elusive hormone as ‘Florigin”. For a long period of time efforts to isolate such a substance have failed. In fact, people have made attempts to collect the substance from the donor plant to a receiver plant through a water jacket, but failed to obtain any stable compound which could induce flowering in other plants. Probably, the suspected florigin may be an extremely unstable, labile and sensitive compound, which could not withstand the most simple extraction methods. However recent experiments involving solvent extraction methods indicate that florigin might be a compound similar to sterols or mRNA-mRNP complex or a labile protein. But there are many plant physiologists who suspect the very existence of such a compound because they feel that some of the known growth promoting hormones by themselves may bring about flower induction by some complex interactions. Most of the known hormones are small molecules, easily transportable and bind to receptors and induce signal transduction. Either such hormones and their binding proteins or the combined complexes may be involved.
However, the transport of such substances has been found to be through sieve tubes, but the rate of translocation in short day plants and long day plants varies. In short day plants, the rate of transportation is about 45-50 cm/hr. but in long day plants, it is about 2-2.5 cm/hr. The rate of translocation of the said flowering substance is found to be 40-100 times slower than the rate of transportation of sucrose, though the components involved in transportation are the same. The different rates of transportation observed in short day plants and long day plants are suspected to be due to the presence of two different substances. It is also known that sieve tubes translocate different substances at different rates because of specific carriers involved in the translocation process. The puzzling feature that is not known is that whether the so called florigin is one compound or a complex of compounds. If it is one compound, then the flowering substance produced by both the short day plants and the long day plants should be the same. If there are two different compounds, the rates of translocation may differ. But why should they differ? The probable explanation is that one of the compounds is constitutively synthesized and such substances reach their destination earlier and the other compound that is synthesized when it is subjected to photo inductive conditions reaches the destination later. However, for the inductive action, both compounds are required, but it is difficult to visualize whether these compounds elicit their action in complexed form or independently at one or two different sites.
But recent studies reveal that plants produce the elusive florigin, which is synthesized in leaves and translocated through sieve cells and reach the base of SAM. The complex of substance now called the actual “Florigin”, is a “Molecule of the century” has been identified as FT (Flowering Time or Flowering locus-T protein correctly Flowering locus T), not the FT mRNA suspected earlier. Actually FT is synthesized in response to constans (CO). It is produced in companion cells of leaves, then it associates with another helper protein and transported to the apex of the stem via sieve tubes. Once it reaches SAM region it combines with another protein called FD, together activate AP1, SOC1 in the apical Meristem, they inturn activate the expression of LFY. Then AP1-LFY triggers the expression of floral homeotic genes. This just explains light induced components, but the flower initiation can also be due to GA, sucrose, vernalization and in a large number of plants it is autonomous. There is a kind of confluence of the products ultimately responsible for triggering the floral homeotic genes.

New model for the photoperiod response in plants; (A) The picture on the left represents the currently accepted model for Arabidopsis, in which light-activated CO overcomes the temperature-dependent inhibition from FLC and induces the expression of FT in the phloem companion cells. FT is moved to the phloem and channeled to the apical meristem where it binds to FD and the complex is recruited into the nucleus. FT–FD binds to the promoter of SOC1 and other meristematic floral integrators, changing the vegetative developmental programme to the ABC programme, eventually producing flowers. (B) The model proposed here includes that depicted in A, but also recruits similar photoperiodic mechanisms to regulate other developmental programme and basic physiological processes. Yellow arrows represent external signals: day/night transition; circadian clock; light quality; and a metabolic signal represented by a fertilizer bottle. Black arrows indicate some of the outputs of the photoperiodic response. http://jxb.oxfordjournals.org/
Red light activated Phy-A induces certain genes, not very well documented, the ultimate flowering inducing product is FT; so also GA has an effect on inducing FT.

Photoperiodic regulation of flowering initiation in Arabidopsis thaliana: Integrating circadian dynamics with physiological processes in plants; Kathleen Greenham; & C. Robertson McClung
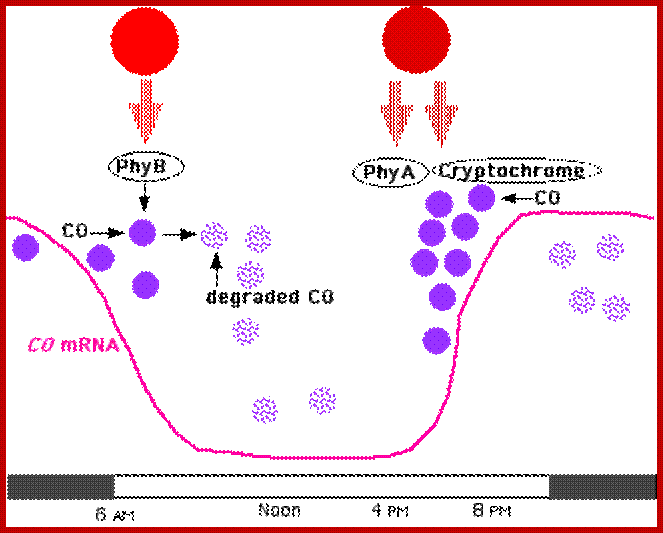
a | Photoperiodic sensing occurs in the phloem companion cells of the leaf. Clock-regulated GIGANTEA (GI) and FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1) activate FLOWERING LOCUS T (FT) transcription through a coherent feed-forward loop, and FT protein (a florigen) in association with another protin is transmitted to the shoot apical meristem (SAM) via the phloem. In the SAM, FT complexes with FD to activate the transcription of several floral meristem identity (FMI) genes, including LEAFY (LFY), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), APETALA1 (AP1) and FRUITFULL (FUL; also known asAGL8). The induction of the FMIs by FT–FD is indirect. FT–FD induces several SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factors, which in turn directly activate the FMIs but which have been omitted for simplicity and clarity. b | GI transcript abundance oscillates owing to transcriptional regulation by the clock, such that on short days GI protein accumulates to a maximum at or near dusk. Although FKF1 transcript and protein abundance also oscillate, the maximum accumulation of FKF1 protein occurs after dusk. Thus, the GI–FKF1 complex accumulates to low levels in the dark and does not include light-activated FKF1. c | Clock-regulated transcription of the CYCLING DOF FACTOR (CDF) genes results in accumulation of CDF mRNA (not shown) and protein in the morning. CDF protein binds to the CONSTANS (CO) promoter to repress transcription. d | Repression of CO transcription is relieved after dusk, but unstable CO protein fails to accumulate. As a consequence, FT transcription and mRNA and protein levels remain low. e | On long days, the peak accumulation of GI protein is phase-delayed and coincides with the peak accumulation of FKF1 protein; both proteins attain maximal levels when it is light, an example of the internal coincidence of two endogenous circadian oscillations. As a result, greater levels of the GI–FKF1 complex accumulate and contain light-activated FKF1. This is an example of external coincidence of light with the endogenous oscillation in FKF1 protein abundance. f | GI–FKF1 complex including light-activated FKF1 degrades CDF proteins, permitting CO transcription, mRNA accumulation and translation of CO protein in the light. g | Light signaling through PHYTOCHROME A (PHYA), CRYPTOCHROME 2 (CRY2) and FKF1 stabilizes CO protein, providing a second example of external coincidence of light, in this case with the endogenous oscillation in CO translation. Stabilized CO protein accumulates and binds to the FT promoter to induce transcription, with subsequent mRNA accumulation. In addition, light-activated FKF1 protein is recruited to the FT promoter to directly stimulate FT transcription and mRNA accumulation. Thus, on long days, FT mRNA abundance increases, permitting translation of FT protein to levels sufficient to induce the transition from vegetative to reproductive growth. Parts b–g are adapted with permission from Ref. (53, Annual Reviews).

Making of the flower; Control of floral meristem identity in Arabidopsis;Trends in Plant Science’ http://www.cell.com/
During the reproductive phase of a plant, shoot meristems follow one of two developmental programs to produce either flowers or vegetative shoots. The decision as to which meristems give rise to flowers, and when they do so, determines the general morphology of an inflorescence. Molecular and genetic research in Arabidopsis and other model species has identified several genes that control the identity that a meristem will adopt. These meristem identity genes are activated in response to developmental and environmental cues, and can be assigned to three basic categories: those required either to initiate or maintain the floral program in some meristems and those required to maintain the vegetative program in others.

Regulation of FM identity; It is regulated by the integration of multiple flowering signals, such as FT and SOC1 perceiving the environmental signals. http://dev.biologists.org/

Pathways regulating the floral transition in Arabidopsis [68,69]. Long photoperiod and gibberellic acids (GAs) promote the floral transition by activating the floral pathway integrators. The enabling pathways regulate floral competence of the meristems by regulating floral repressor activity such as the FLC. Arrows indicate activation and short lines ending with a dot indicate repression. The data underlying the model and the corresponding homologs in citrus are presented in Table 1. A plus sign (+) indicates up-regulation and a minus sign (-) indicates down-regulation in precocious trifoliate orange. Genes in red type were not found by MPSS. http://www.biomedcentral.com/

All the above mentioned pathways integrate and converge of Floral Meristem (FM) identity genes; which direct the meristem to initiate flowering. www.cell.com.
Most angiosperm flowers are tightly integrated, functionally bisexual shoots that have carpels with enclosed ovules. Flowering plants evolved from within the gymnosperms, which lack this combination of innovations. Paradoxically, phylogenetic reconstructions suggest that the flowering plant lineage substantially pre-dates the evolution of flowers themselves. We provide a model based on known gene regulatory networks whereby positive selection on a single, partially redundant gene duplicate ‘trapped’ the ancestors of flower-bearing plants into the condensed, bisexual state ∼130 million years ago. The LEAFY (LFY) gene of Arabidopsis encodes a master regulator that functions as the main conduit of environmental signals to the reproductive developmental program. We directly link the elimination of one LFY paralog, pleiotropically maintained in gymnosperms, to the sudden appearance of flowers in the fossil record.

http://www.mun.ca/

www.plantphys.info


A simple diagram showing the four major genetic pathways regulating flowering time in Arabidopsis. photoperiodic, light quality, vernalization, autonomous and GA pathways.The two main pathways mediating environmental responses are the long‐day and vernalization pathways. The two pathways thought to function independently of environmental cues are the autonomous pathway, which promotes flowering in all conditions, and the GA pathway, which is needed for flowering in non‐inductive short‐day conditions. http://embor.embopress.org/
Effect of GA on Flowering:
Fascinating aspect of flower inducing substance (s) is that when the extracts, obtained from photo induced leaves of Xanthium is applied to lemma plant kept under non inductive conditions, the extracts induce flowering in lemma plants. However, the induction of flowering by the extracts should be supplemented with gibberellins, without which the extracts alone or gibberellins alone has no effect. This experiment suggests that gibberellins are probably one of the components of elusive florigin (like elusive “Himalayan snow man’), but the nature of the other component is still a mystery.
It is very well known that gibberellins induce bolting and flowering in rosette leaved long day plants, but not in short day plants (with some exceptions). In long day plants, GA not only stimulates the elongation the condensed internodes, but at the same time, it also promotes the formation of factors needed for flowering. Thus GA treatment substitutes photoperiodic treatment in long day plants. In addition to this complexity, application of high concentration of Gibberellins and Cytokinins to the callus, obtained from Arachis hypogea (peanut plant), resulted in the induction of flowers directly from the callus. Paradoxically ABA, a growth inhibitor, is very effective in inducing flowers in some short day plants like Fragilis, Pharbatis, etc. But ABA does not induced flowers in xanthium, another short day plant. This particular case is very interesting, at the same time, it is also intriguing and it further raises doubts about the existence of florigin per se. But such experimental results are very few and conditions used for such materials have to be carefully analyzed. Much more perplexing that is observed in some cases is that the application of cytokinins to the whole plant induces flowering, when the plant is at a particular stage of development.
Based on gibberellin’s promotive effect on long day plants and its failure on short day plants, Brain and his colleagues (1958-59) came out with a working model to explain the action of photoperiods on flower inducing substances. According to this model, during photo inductive red light treatment a precursor gets converted to Gibberellin or Gibberellin like substance. The same substance is believed to undergo decaying back to the precursor either in dark or under far-red light treatment. According to their concept it is assumed that Gibberellin like substances have to be maintained at high concentrations in long day plants to be effective in producing the elusive compound called ’florigin’. But in short day plants, according to Brain, et.al. GA like substances are effective only when their concentration is very low for higher concentration of GA is inhibitory. But the said scheme of events fails to explain how low levels of GA like substances can produce sufficient quantities of florigin in short day plant to bring about the induction of flowers and it is expected that the florigin that is produced in short plants or long day plants should be the same.
Even today, with all the knowledge of molecular biology of the exact processes involved in inducing flowers are not known. Still, it is very important to understand the model proposed by Chailakhyan (1936), a Great Russian plant scientist. He toiled his entire life time to understand this phenomenon and his proposed model is very worth understanding. Cajlachjan, another way to pronounce his name, has assumed that the florigin formation takes place at two levels but in two steps. Further, florigin is not one substance but it is a complex of two substances, i.e., gibberellins and Anthesins. It is also assumed that long day plants synthesize anthesins irrespective of photoperiods, which means anthesins are produced constitutively, which indicates that the genes responsible for the synthesis of the above said compound are constitutively expressed all the time in long day plants. But the synthesis of GA or GA like compounds is under the control of long day photoperiodic conditions. In the sense, the pFR produced in long day plants has an important role in activating the pathway of GA synthesis. On the other hand, short day plants are believed to synthesize GA constitutively and the synthesis of anthesins is under the control of short day photoperiods. It means the pR form of the pigments is effective in inducing the synthesis of anthesin.
Gibberellin pathway and Light quality pathway. Each of them has certain products and they get integrated at the base of the meristem and determine and differentiate the floral meristem. Many of the components that are synthesized in floral meristem or axis, very young leaf primordials ultimately interact and integrate in activating the apical Meristem into floral meristem.

The nature of floral signals in Arabidopsis; Summary of findings here and in the companion paper of positive effects (arrows) on flowering and CO/FT for two commonly used LD photoresponses. This schematic incorporates effects on FT and flowering of: mutants; gene silencing; change in light intensity; and a block to photosynthesis. Predominantly, in LD, photosynthetic sucrose amplifies CO/FT expression (see companion paper) while phytochrome acts directly and also via GA, which plays a permissive and, often, non-limiting role. There is also a direct but lesser LD-mediated increase in GA supply via the petiole response to FR-rich light. A dashed arrow indicates a potential step of regulation, and weaker responses are indicated by thinner arrows. The electronics symbol for a speaker is used to show sucrose amplification of CO/FT expression. http://jxb.oxfordjournals.org/

http://journal.frontiersin.org/
Light has another discerning, but subtle activity is inducing the synthesis of GA1 and its derivatives that activate LFY, and hypocotyl elongation and shade avoidance processes.
In spite of numerous experiments and detailed studies well over 70 years or so, the explanation given to describe the entire mechanism of flower induction is confusing and often contradictory in nature. Particularly investigations at the molecular level are few and they are not convincing. Skoog and his students working on a cultivar variety of Nicotiana tabbacum plant demonstrated that when the total DNA extracted from the leaves of photo induced plants is applied to the stem tips of tobacco plants kept under non photo inductive conditions, the receiver plant flowered. It is not clear from their experiments whether on not the DNA extracted is really pure and free from proteins and other stimulatory small molecules that can easily diffuse into stem apex. Yet it indicates that pure DNA perse cannot express until and unless they are provided with regulatory factors which are mostly proteins. If proteins are associated with such extracted DNA, then what are those proteins? Unfortunately, nothing is known about them. But it is clear from their experiments that the applied DNA is not free from proteins and such proteins are highly stable when they are associated with DNA. It would be interesting to isolate chromosomal proteins from induced plant and provide the same to non induced plants and see whether the receiver plants produce flowers or not.
A group of Japanese workers demonstrated an increase in the levels of different species of RNAs and different species of proteins is buds during floral initiation. Unfortunately it is not clear whether there are any qualitative difference among the mRNAs and proteins produced during floral initiation. It is very important to understand the molecular events that lead to the transformation of vegetative buds into floral buds. It beats any one’s imagination why such work has not been successfully elucidated so far; even though the technology is available at hand.
Nevertheless, it is now clear that the induction of flowering due to photoperiodic effects is regulated at two sites separated by space and in time. In the first event the site of response to photoperiodic induction is young leaves. The second site is the terminal or axillary shoot buds. Which on receiving the flower inducing substance(s) are stimulated to produce floral axis or floral buds? Shoot buds per-se do not respond to photo inductive treatments, but they respond when leaves are subjected to such treatment. Which means whatever substances produced in leaves reach the vegetative buds and induct flowering in them?
At the level of leaves, phytochromes able to receive signals from proper photoperiodic treatments, undergo excitation and induce gene expression in the cells leading to the synthesis of flower inducing substances. According to the prevailing views, the pFR pigments produced in long day plants induce the synthesis of GA or Gibberellin like compounds in proplastids/plastids and the same are released into cytoplasm. In the same long day plants, whatever small amounts of pR pigment found in the leaf cells might be able to induce gene expression to produce anthesins all the time irrespective of photoperiodic treatment. On the contrary in short day plants, the pR form of pigments that accumulated during long dark periods are suppose to induce anthesins production and the little amount of pFR present might be involved in producing GA like compounds constitutively all the time. The anthesins appears to be proteinaceous or protein likes compounds.
That is why the rate of translocation of flowering compounds varies. Anthesins appears to be translocated faster than gibberellins. Whether the genes that respond to two different photoperiodic treatments are located in the nucleus or plastids or both, is difficult to visualize, until and unless one obtains nuclear or cytoplasmic mutants for flowering.
Once such components are produced in leaves they are translocated to different site of cell vegetative buds. It is conceivable that as long day plants produce anthesins and short day plants produce GA like compounds constitutively; they reach the meristems of the vegetative buds earlier. On the contrary as GA like substances in long day plants and in short day plants produced under photo inductive conditions; reach the vegetative mounds later. If one of the components reaches the vegetative meristems earlier, it cannot bring about the total gene expression without the presence of the other factor. So both the factors are required in the vegetative buds for total and comprehensive effect to be effective. Once the vegetative buds receive both factors (i.e. GA and Anthesins) in required quantities, the florigin complex activates a battery of genes required for the transformation of vegetative buds into floral buds.
Whether the florigin components activate one gene or a group of genes, the activation leads to a cascade effects. It means a specific gene or gene products may activate group or group of genes, which inturn may activate another set of genes. The products of these genes ultimately bring about the transformation of vegetative meristems into floral meristems and commit them to develop into floral structures. It is important to remember that the production of stalk, bracts, sepals, petals, stamens and pistils is not a single event nor is it regulated by a single gene. They are a series of sequential events which require a sequential expressions and gene products. The totality of this highly regulated gene expression results in the transformation of vegetative buds into highly organized floral buds of a particular type. As many plants belonging to different species produce flower where all are sepals, some lacking petals, stamens or pistils or the combination of any of them. It is predictable that the development of each of these structures is under the control of specific genes or gene clusters.
It is to be remembered that the molecular biology of flower induction and development is in progress and we have to wait the D-day for understanding the whole process in Toto.
Phytochrome Pfr activated gets autophosphorylated. It can act on membrane G protein receptor that can lead to cGMP and calcium modulated Cam, both can enter into the nucleus. What is their function is not clear. Activated PhyB-Pfr complex is a serine-threonine protein kinase. Once it enters into the nucleus it interacts and binds to PIF3 dimeric (phytochrome interacting factor) proteins and also binds to HFR1-PIF3 dimers (HFR1 = long hypocotyl far-red protein1 is a TF); then PhyB-p and receptor proteins bind to their respective gene regulatory elements called G-box receptor and recruit TFs and RNAP II and activate genes like CCA (TF), LHY (TF) and HY5 (TF). These gene products perhaps activate transcription of FT in association with Constans (CO) in leaf cells. Probably they activate FT gene in the vascular bundles, especially companion cells of leaves, where the role of PhyB plays in the production of FT.
Vernalization pathway:
Many plants have to go through wintering to flower in the next season. The famous plant is petkus rye, which is essential for bread makers in Russia. It is not the only plant that requires cold treatment for the plant to flower. These plants, particularly seeds contain specific proteins and its associated component which bind to loci that are involved flower induction and silence the chromatin by heterochromatization. Heterochromatization is achieved by histone methylation and histone deacetylation at specific loci. One such protein that is bound to chromatin is known as flowering locus C (FLC). FLC acts as a repressor. Cold treatment in fact induces few FLC antagonizing genes and FLC dissociates from the loci and get degraded or remain free. There are several genes that are involved are VRN1, VRN2, VRN3 and VIN3. Even Frigida (FRI) proteins are involved. Frigida promotes FRI antagonizes FLC. Once the chromatin is free from FLC and its associated proteins, they interact with floral integrators such as SOC, CO, FT and LFY, which inturn activate genes for floral parts. An important event that takes place is the movement of Flowering locus T ( FT) translocate from leaves to SAM In certain cases GA can overcome vernalization.

Pathways controlling flowering-time inArabidopsis. The flowering-time pathways control the expression of the floral pathway integrators SUPPRESSOR OF OVEREXPRESSION OF (CAL) and LFY, which convert the vegetative meristem to a floral fate. Recent expression data has indicated that FUL may also act as a floral integrator (Schmid et al., 2004). The photoperiod, gibberellin, light-quality and ambient-temperature pathways activate floral pathway integrators. The CONSTANS (CO) transcription factor functions in the photoperiod pathway; long-day photoperiods promote flowering by circadian clock (CLOCK) dependent and independent mechanisms, which control the activity of CO. Activation of flowering is antagonized by the floral repressors encoded by (shown in green) FLOWERING LOCUS C (FLC), FLOWERINGLOCUSM (FLM), TERMINALFLOWER1 (TFL1), TERMINAL FLOWER2 (TFL2), SHORT VEGETATIVE PHASE (SVP), TARGET OF EAT1 (TOE1), TARGET OF EAT2(TOE2), SCHNARCHZAPFEN (SNZ), SCHLAFMUTZE (SMZ)and EMBRYONICFLOWER1/2 (EMF1, EMF2). TFL1 may also be downstream of CO, as it is induced after CO activation (Simon et al., 1996). FLC expression is controlled by a number of different pathways.
The genes shown in purple, FRIGIDA (FRI), FRIGIDA-LIKE1(FRL1), FRIGIDA-LIKE2 (FRL2), PHOTOPERIOD INSENSITIVE EARLY FLOWERING1 (PIE1), AERIAL ROSETTE1 (ART1), EARLY UNDER SHORT DAYS4 (ESD4), VERNALIZATION INDEPENDENCE3 (VIP3) and VERNALIZATION INDEPENDENCE4 (VIP4), encode proteins that promote FLC expression and delay flowering. FLC expression is down regulated in response to prolonged cold by proteins encoded by the genes (shown in blue) VERNALIZATION INSENSITIVE3 (VIN3), VERNALIZATION1 (VRN1) and VERNALIZATION2 (VRN2), and also by proteins encoded by the genes of the autonomous pathway (red): FCA, FY, LUMINIDEPENDENS (LD), FLOWERING LOCUS D (FLD), FVE, FLOWERING LOCUS K (FLK) and FPA. The distinction between potential transcriptional and post-transcriptional functions of genes of the autonomous pathway is not made here, but is shown more clearly in Fig. CONSTANS1 (SOC1), FT and LEAFY (LFY). These genes encode proteins that activate the floral meristem identity (FMI) genesAPETALA1 (AP1), APETALA2 (AP2), FRUITFULL (FUL), CAULIFLOWER; www.dev.biologists.org

The Photoperiod and FLC Pathways Interact in the Floral Transition:In favorable photoperiods, CO activates FT in the leaf veins, and this leads to the induction of flowering. FT RNA or FT protein moves in the phloem to the shoot apex, where its interaction partner FD is expressed. It is unclear how FT moves from the end of the vasculature into the meristem. At the shoot apex, FT and FD together activate AP1. FLC represses the floral transition by antagonizing FT upregulation in the leaf veins and FD and SOC1expression at the shoot apex. AP1 and SOC1 induce flowering; Isabel Bäurle1, and Caroline Dean, http://www.cell.com/

MADS genes control Flowering; A model for the regulation of BM5 expression by vernalization and spring habit genotypes. Vernalization controls two genetic systems that regulate BM5 transcription through the BM5 promoter. One represses BM5 expression in plants that have not been vernalized. Extended cold treatment counteracts this repression and activates a second regulatory mechanism that activates BM5transcription. Known flowering time genes such as the barley equivalent of SOC1 are likely to be involved in this activation pathway. Recessive spring habit alleles of the Vrn-H2 locus abolish the repression pathway and allow some expression of BM5. Dominant alleles of Vrn-H1 activate expression regardless of vernalization status, but do not completely counteract repression, leading to some BM5activity. Combining both spring habit genotypes results in BM5 expression levels that approach those in vernalized plants, through simultaneous activation and de-repression of the BM5 promoter. www.pnas.org
Cold has positive effect on the activation of genes such as BMS genes that leads to floral transition.

Control of Arabidopsis flowering: the chill before the bloom; During vegetative growth FLC gene is expressed controlled by FRL1,2,ESD4,ART1,PIE1, VIP3 and VIP4; the said proteins maintain FLC protein in active state. In autonomous pathway functions antagonistically against the genes that express FLC. Some of the genes are FCA,FPA and FLK and Polyadenylation factor FY function post transcriptionally. The FVE/FLD proteins function post transcriptionally act as histone deacetylase (HDAC) promote FLC into inactive state. The FLC can also be repressed by long periods of vernalization (exposure to cold). Prolonged cold induces the expression of VIN3 which promotes an inactive FLKC chromatin state, then VRN1 and VRN2 are recruited to FLC and perform Histone methylase, thus causing repression of FLC; http://dev.biologists.org/
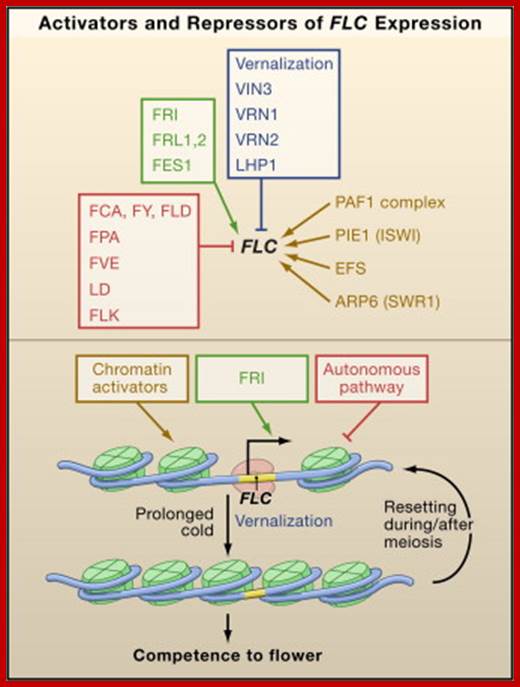
Regulation of FLC Expression: (Top panel) Expression of the floral repressor FLC is activated and repressed by many pathways. The autonomous pathway (red) represses FLC expression and comprises FCA, FY, FLD, FPA, FVE, LD, and FLK. Vernalization represses FLC through VIN3, VRN1, VRN2, and LHP1 (blue). FRI is a strong activator of FLC expression and acts together with FRL1, FRL2, and FES1 (green). Several chromatin regulators (brown) activate FLC expression (PAF1 complex, PIE1, EFS, and ARP6).
(Bottom panel) Regulation of FLC chromatin structure. FLCactivators promote an active chromatin state at FLC, whereas many components of the autonomous pathway promote an inactive chromatin state. Vernalization causes remodeling of FLC chromatin and accumulation of histone modifications characteristic of heterochromatin. During or after meiosis, high FLC expression is reset to restore the vernalization requirement in the next generation; www.cell.com

Deacetylation leads to progressive silencing of FLC during cold; and stable repression in subsequent growth by methylation of gene histone tails; Plant cell Biol

The Arabidopsis genome sequence was completed in 2000. This has led to a great increase in our understanding of the molecular basis of both plant development and the response to environmental stimuli. Having the genome sequence has enabled genomics approaches which aim to assign a function to each of the predicted 26,000 genes. Knockout mutations generated by insertional mutagenesis or gene silencing with RNAi methodology suggest a function for a gene if the mutants can be linked to a phenotype. Oligonucleotides corresponding to each of the predicted genes can be spotted on a microarray which can be used to determine the pattern of expression of each of the genes, again, suggesting a function. The knowledge of gene function is deepening our understanding of development in Arabidopsis, and importantly, it is enabling Arabidopsis to be a platform into similar levels of understanding in our major crop plants.www.cropscience.org

Family of VERNALIN like gene product
Vernalization leads to expression of specific Genes:
Many plants have to go through wintering to flower in the next season. The famous plant is petkus rye, which is essential for bread makers in Russia. It is not the only plant that requires cold treatment for the plant to flower. These plants, particularly seeds contain specific proteins and its associated component which bind to loci that are involved flower induction and silence the chromatin by heterochromatization. Heterochromatization is achieved by histone methylation and histone deacetylation at specific loci. One such protein is known as flowering locus C (FLC). FLC acts as a repressor. Cold treatment in fact induces few FLC antagonizing genes and FLC dissociates from the loci and get degraded or remain free. There are several genes that are involved are VRN1, VRN2, VRN3 and VIN3. Even Frigida (FRI) proteins are involved. Frigida promotes FLC and FRI antagonizes FLC. Once the chromatin is free from FLA and its associated protein, they interact with floral integrators such as SOC, CO, FT and LFY, which inturn activate genes for floral parts. In certain cases GA can overcome vernalization.

Microarray of gene expressed products during induction pathway

A simple diagram showing the four major genetic pathways regulating flowering time in Arabidopsis. The two main pathways mediating environmental responses are the long‐day and vernalization pathways. The two pathways thought to function independently of environmental cues are the autonomous pathway, which promotes flowering in all conditions, and the GA pathway, which is needed for flowering in non‐inductive short‐day conditions.; Flowering on time; embor.embopress.org
Genetics of Flowering:
Gregor John Mendel showed that the color of the flower is controlled by the action of a pair of heritable factors, now called Genes. They are independent discrete units of heredity. Inductive stimulus should transform the entire shoot apical meristem (SAM) into flower inducing meristem; there again from the same meristem different organs like sepals, petals; stamens and ovary have to develop; all of them are transformed laves. The flower is indeed is a modified shoot with leaves representing different parts of the flower. Stamen and ovary differentiation is more complex than sepals and petals for the simple reason they have highly specialized structures and they are gamete producing super-specialized reproductive organs.
Whether it is light induced pathway or vernalization pathway or GA induced pathway or autonomous pathway, signals are produced and translocated to phloem and to the base of SAM. The important component that induces FT is CO which activates the expression of FT which is a 20Kda protein. The FT in association with FD (TF) activates AP1 and LFY; this leads to the activation floral organ genes.

Photoperiodic control of flowering; Trends in plant science; www.cell.com; Photoperiodic control of flowering; The timing of floral transition has a direct impact on reproductive success. One of the most important environmental factors that affect the transition is the change in day length (photoperiod). Classical experiments imply that plants monitor photoperiods in the leaf, and transmit that information coded within an elusive signal dubbed florigen to the apex to reprogram development. Recent advances in Arabidopsis research indicate that the core of the day-length measurement mechanism lies in the circadian regulation of CONSTANS (CO) expression and the subsequent photoperiodic induction of the expression of FLOWERING LOCUS T (FT) gene, which might encode a major component of florigen. In this review, we introduce current perspectives on how, when and where the floral signal is generated. Trends in Plant Science; http://www.sciencedirect.com/; http://www.cell.com/

Multiple developmental pathways for flowering in Arabidopsis:(a) the photoperiodic (long day) pathway, which operates in the leaves; (b) the convergent autonomous (leaf number)/vernalization (low temperature) pathway; (c) the carbohydrate (sucrose) pathway; and (d) the gibberellin pathway. Fonte: In Arabidopsis, FT binds to FD, and the FT/FD protein complex activates the AP1 (Apetala1) andSOC1 genes (suppressor of over expression of CO1), which trigger the LFY (Leafy) gene expression.LFY and AP1 then trigger the expression of the floral homeotic genes. The autonomous (leaf number) and vernalization (low temperature) pathways act in the apical meristem to negatively regulate FLC-flowering locus C, a negative regulator of SOC1. The sucrose and gibberellin pathways, also located in the meristem, promote SOC1 expression; www.intechopen.com
The above figure is self illustrative and shows how LD and SD effect the activation of gene, and the products move into the sieve cells, from there they reach the SAM. Note Hd3a is a homolog of FT in rice plants
Once, FT and FD act together on AP1 and LFY floral Meristem evocates and the floral organs start developing. Each of the structures is controlled by single or a combination of genes. The famous ABC model has been used to explain the genes involved. Mutants in said gene exhibit their phenotypic characters. The gene-A produces sepals and combination of AB generates petals and the combination of B and C generate stamens and C alone produces Carpels. As stamens and carpels contain different set of structures, to explain them the ABC model has be extended as ABCD and E model.

Flowering time regulation; Photoperiod and temperature sensing in Leaves; Young Hun Song et al;www.cell.com



Floral induction and determination; where is flowering controlled? http://www.cell.com/
The ABC Model of Flower Development; http://users.rcn.com/
Genetic analysis of mutants — especially those found in the dicots Arabidopsis thaliana and in the snapdragon (Antirrhinum) support the ABC model of flowering. This model postulates a group of genes that encode the transcription factors needed to turn on the genes for sepal, petal, etc. development. The "master switches" fall into 3 groups: A, B, and C. Following are the rules;
- Cells in which only A genes are expressed develop into sepals. This occurs at the lowest level of the floral meristem.
- Cells in which both A and B genes are expressed develop into petals. This occurs at the next higher level.
- Expression of B and C genes turns on the developmental program to form stamens.
- Expression of C genes alone turns on the development of carpels in the innermost band of cells.
- Examples of A, B, and C group genes involved in
flowering.
These have been identified in Arabidopsis thaliana. A group APETALA1 (AP1) and
APETALA2 (AP2)B group APETALA3 (AP3) and
PISTILLATA (PI)C group AGAMOUS (AG)
The transcription factor LEAFY plays a major role in turning on the A, B, and C group genes in the appropriate locations:
- The LEAFY protein alone turns on AP1 in whorls 1 and 2.
- LEAFY plus a protein encoded by the gene UFO (for "unusual floral organs") turn on AP3 in whorls 2 and 3.
- LEAFY and a second, still unidentified, protein turn on AG in whorls 3 and 4.
If LEAFY protein alone is sufficient to turn on AP1, why isn't AP1 expressed in all four whorls?
The answer: AGAMOUS blocks the expression of AP1, so any cell expressing AGAMOUS cannot express AP1.
In fact, the antagonism is reciprocal: AP2 in whorls 1 and 2 (A group) inhibits AGAMOUS so the gene expression in whorls 3 and 4 remains distinct from that in whorls 1 and 2.
The proteins encoded by APETALA3 and PISTILLATA (Group B) form a heterodimer that binds to
- the APETALA1 protein to form petals
- the AGAMOUS protein to form stamens.
Aided by a fourth transcription factor encoded by the gene SEPALLATA3, these quaternary complexes bind to specific sequences of DNA turning on the expression of the various genes needed to form whorls 2 and 3. Further research may reveal similar behavior for the other genes.
|
SEPALLATA3 (SEP3) is one of four SEP genes in Arabidopsis. If all but SEP4 are inactivated, a flower with only sepals is formed (hence the name). If all four are inactivated, no flowers are formed at all. |
So formation of a flower requires a cascade of sequential gene activity that gradually converts a mass of undifferentiated cells (the apical meristem) into the parts of a flower. The genes encode transcription factors that act as master switches, turning on (or off) downstream genes that ultimately make each part of the flower in its appropriate location.
This same strategy of genetic control of developmental pathways is seen in animal development. Try this link to see some examples in Drosophila.
Recommended reading: The Genetics of Flower Development by Elliot M. Meyerowitz (in whose lab many of these discoveries were made). It was published in the November 1994 issue of Scientific American.

www.users.rcn.com



What is interesting thing about these genes is the said gene products are a group of MADS genes and they are homeotic genes. Each of these have been identified and studied.

MADS box proteins in combination induce specific floral organs. Look at A, AB, CD and D alone and the pervasive E overlaps ABCD. MADS are mostly transcription factors and they act in combination with one another. They have many domains from N to C end.


MADS=MCM, Agamous, Dificiens and SRF

Ap3 expression with Gus as a reporter gene in apical meristems destined to
Become floral organs.

The figure is the repeat- demonstrates the inputs for the floral organogenesis.
This figure is an ultimate- start to and finishing of flowering, starts at leaves and ends in flowers.
I have taken few authors written articles and put them together, so student can understand experts views on flowering. I duely acknowdge the authors for their great insight into the the most complex flowering process.
Jan A. D. Zeevaart, MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI; September, 2007
Introduction
Florigen as a Physiological Concept:
Specific flower-inducing substances were first postulated by Julius Sachs (1865), but more convincing evidence had to await the discovery of photoperiodism (Garner and Allard 1920). The seminal finding was that in photo periodically sensitive plants the day length is perceived by the leaves, whereas flower formation takes place in the shoot apical meristem (Knott 1934). This finding demonstrates that a long-distance signal, called the floral stimulus or florigin (Chailkhyan 1936), moves from an induced leaf to the shoot apex. The floral stimulus can be transmitted from a flowering partner (donor) via a graft union to a non-flowering partner (receptor), as illustrated in Figure 25.29 of the textbook. Furthermore, it was shown that florigen is exchangeable between related species and genera, as well as among different photoperiodic response types (Lang 1965; Zeevaart 1976; Zeevaart 2006). These observations led to the hypothesis that florigen is wide-spread, if not universal, in flowering plants, and that only the conditions that regulate its production vary among the different response types. Although certain physiological characteristics of florigen, such as its movement in the phloem (e.g., King and Zeevaart 1973), could be investigated, its identity remained unknown. Thus, florigen remained a physiological concept rather than a chemical entity.
With the isolation of auxin as the first-identified plant growth hormone in the 1930s and the discoveries of cytokinins and gibberellins in the 1950s, many attempts were made to extract florigin. In the early approaches, it was assumed that like the classical plant hormones, florigen would be a small organic molecule. Extracts prepared from flowering material were tested for flower-promoting activity in vegetative plants. Positive results were reported occasionally, but none of them was reproducible (reviewed in Zeevaart 1976). As a result, skeptics challenged the adequacy of florigin as a single substance causing flowering. Instead, it was proposed that florigen consists of multiple factors and that flowering is induced by a specific ratio of known hormones and metabolites (Bernier 1988; Bernier et al. 1993).
The genetic network of floral transition at the shoot apical meristem; The genetic basis of flowering responses to seasonal cues; Fernando Andrés & George cCupland;

![Description: <p>Figure 2. FT fulfills the criteria for the flowering hormone Florigen.<i> FT</i> and its closest relative <i>TSF</i> are expressed in the leaf veins from where the protein moves to the shoot apex. The proteins interact directly with the transcription factor FD and trigger transcriptional reprogramming that switches the vegetative apical meristem into an inflorescence meristem [4]. Expression of <i>FT</i> is stronger than that of <i>TSF</i> explaining that the effect of a mutation in the<i> TSF</i> gene is best visible in a <i>ft</i> mutant background.</p>](Plant_Growth_And_Development10-Physiology_Of_Flowering_files/image124.gif)
FD-flowering locusD dark blue and Light blue FT; http://www.nature.com/Genetics


Gene networks controlling the initiation of flower development; ;http://www.cell.com/

A Simplified Model of Florigen Cycling and Transport in a “short-night” (Arabidopsis) versus a “long-night” (rice) plant. Proteins (red) include: FT (Flowering Locus T), Hd3a (Heading date 3a), CO (Constans) , HD1 (Heading date 1), and Ehd1 (Early heading date 1). Proposed florigens inArabidopsis (FT) and rice (Hd3a) cycle in leaves with circadian rhythm, but with different phases in response to different photoperiods. At dawn, when photosynthesis resumes in leaves, phloem transport from leaves to SAMs also resumes, carrying florigen. If florigen is above a threshold level, then it may trigger the floral transition in the SAM. http://www.howplantswork.com/
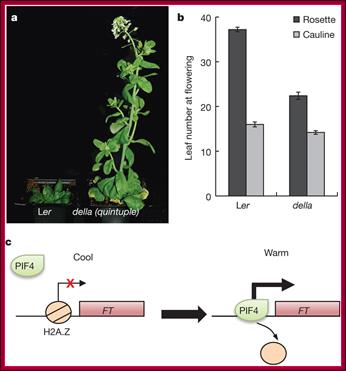
www.pcb.org

Regulation of Arabidopsis thaliana FT transcription; The genetic basis of flowering responses to seasonal clues; Transcription of FLOWERING LOCUS T (FT) is a point of convergence of different seasonal cues and is tightly regulated. A 5.7 kb region 5′ of the ATG represents the FT promoter and contains all of the elements that are required to mediate spatial and temporal expression of FT in response to photoperiod. CONSTANS (CO) activates FT transcription in response to long days and was recently demonstrated to bind in vitro and in vivo to the proximal region of the FT promoter. Interestingly, although CO binds to the proximal region of the promoter close to the transcription start site, a more distal region (shown by the dotted arrow in the figure) also seems to be essential for CONSTANS (CO)-dependent FT activation. This upstream region might be recognized by an unknown activator complex that enhances the affinity of CO to bind to the region located in the proximal promoter or is required for CO function. The CCAAT-box-binding complex has been proposed to carry out such a function. GIGANTEA (GI), FLAVIN KELCH F BOX 1 (FKF1) and CYCLIC DOF FACTOR 1 (CDF1), which affect expression of FT by regulating CO mRNA level, are also reported to bind directly to the FT locus. Indeed, tagged CDF1 proteins expressed in the fkf1 mutant strongly associate near the FT transcription start site, suggesting that FKF1 contributes to FT transcriptional activation by removing CDF repressors from its locus. The basic helix–loop–helix transcription factor PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) binds directly to the proximal FT promoter in response to high temperature when it activates FT transcription independently of CO, and another basic helix–loop–helix transcription factor, CIB1, binds to a similar region and activates FT in response to blue light109. LIKE HETEROCHROMATIN PROTEIN1 (LHP1) is a direct repressor of FT transcription. In lhp1 mutants, FT mRNA is highly expressed, and flowering is accelerated independently of photoperiod110. LHP1 is a chromo domain protein that colocalizes with histone H3 modified by trimethylation on lysine 27 (H3K27me3), which is a repressive chromatin mark formed by Polycomb repressive complex 2 (Ref. 111). LHP1 is proposed to be a plant-specific component of Polycomb repressive complex 1 (Ref. 111), chromatin immunoprecipitation (ChIP) experiments (such as ChIP followed by microarray (ChIP–chip) and ChIP followed by high-throughput sequencing (ChIP–seq)) showed the pattern of accumulation of these repressive marks along the locus of FT (upper panel of the figure). Both H3K27me3 and LHP1 widely cover the FT locus, conferring its transcriptional repression, whereas those regions that are free of repressive marks have been proposed to constitute a window of open chromatin that is accessible to regulatory factors104. Many of the other proteins that bind the FT locus are also transcriptional repressors. The MADS box proteins SHORT VEGETATIVE PHASE (SVP) and FLOWERING LOCUS C(FLC) confer vernalization requirement and repress FT by binding CArG boxes present in its promoter and first intron. SVP and FLC are proposed to bind FT in a heteromeric complex. The AP2-like transcription factor TEMPRANILLO (TEM) genes TEM1 and TEM2 also repress FT expression. Moreover, TEM1 binds directly to the 5′ untranslated region of FT. A balance between CO and TEM genes has been described as an important mechanism in the transcriptional control of FT particularly in young plants113. Other repressors of FT belong to a small group of AP2-like genes, mRNAs of which are targets of miR172. This group includes APETALA2 (AP2), TARGET OF EAT 1 (TOE1), TOE2, SCHNARCHZAPFEN (SNZ) and SCHLAFMÜTZE (SMZ). However, only SMZ is known to bind directly to a region 1.5 kb downstream of the FT stop codon. Fernando Andrés & George Coupland; www.natrure.com
The transition to flowering is a major developmental switch in the life cycle of plants. In Arabidopsis (Arabidopsis thaliana), chromatin mechanisms play critical roles in flowering-time regulation through the expression control of key flowering-regulatory genes. Various conserved chromatin modifiers, plant-specific factors, and long noncoding RNAs are involved in chromatin regulation of FLOWERING LOCUS C (FLC, a potent floral repressor). The well-studied FLC regulation has provided a paradigm for chromatin-based control of other developmental genes. In addition, chromatin modification plays an important role in the regulation of FLOWERING LOCUS T (FT, encoding Florigen), which is widely conserved in angiosperm species. The chromatin mechanisms underlying FT regulation in Arabidopsis are likely involved in the regulation of FT relatives and, therefore, flowering-time control in other plants. http://www.cell.com/trends/
Regulation of Arabidopsis thaliana- FT transcription;
Transcription of FLOWERING LOCUS T (FT) is a point of convergence of different seasonal cues and is tightly regulated. A 5.7 kb region 5′ of the ATG represents the FT promoter and contains all of the elements that are required to mediate spatial and temporal expression of FT in response to photoperiod. CONSTANS (CO) activates FT transcription in response to long days and was recently demonstrated to bind in vitro and in vivo to the proximal region of the FT promoter. Interestingly, although CO binds to the proximal region of the promoter close to the transcription start site, a more distal region (shown by the dotted arrow in the figure) also seems to be essential for CONSTANS (CO)-dependent FT activation. This upstream region might be recognized by an unknown activator complex that enhances the affinity of CO to bind to the region located in the proximal promoter or is required for CO function. The CCAAT-box-binding complex has been proposed to carry out such a function. GIGANTEA (GI), FLAVIN KELCH F BOX 1 (FKF1) and CYCLIC DOF FACTOR 1 (CDF1), which affect expression of FT by regulating CO mRNA level, are also reported to bind directly to the FT locus. Indeed, tagged CDF1 proteins expressed in the fkf1 mutant strongly associate near the FT transcription start site, suggesting that FKF1 contributes to FT transcriptional activation by removing CDF repressors from its locus26. The basic helix–loop–helix transcription factor PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) binds directly to the proximal FT promoter in response to high temperature when it activates FT transcription independently of CO, and another basic helix–loop–helix transcription factor, CIB1, binds to a similar region and activates FT in response to blue light. LIKE HETEROCHROMATIN PROTEIN1 (LHP1) is a direct repressor of FT transcription. In lhp1 mutants, FT mRNA is highly expressed, and flowering is accelerated independently of photoperiod. LHP1 is a chromo domain protein that colocalizes with histone H3 modified by trimethylation on lysine 27 (H3K27me3), which is a repressive chromatin mark formed by Polycomb repressive complex 2 . LHP1 is proposed to be a plant-specific component of Polycomb repressive complex 1 , chromatin immunoprecipitation (ChIP) experiments (such as ChIP followed by microarray (ChIP–chip) and ChIP followed by high-throughput sequencing (ChIP–seq)) showed the pattern of accumulation of these repressive marks along the locus of FT (upper panel of the figure). Both H3K27me3 and LHP1 widely cover the FT locus, conferring its transcriptional repression, whereas those regions that are free of repressive marks have been proposed to constitute a window of open chromatin that is accessible to regulatory factors. Many of the other proteins that bind the FT locus are also transcriptional repressors. The MADS box proteins SHORT VEGETATIVE PHASE (SVP) and FLOWERING LOCUS C(FLC) confer vernalization requirement and repress FT by binding CArG boxes present in its promoter and first intron. SVP and FLC are proposed to bind FT in a heteromeric complex. The AP2-like transcription factor TEMPRANILLO (TEM) genes TEM1 and TEM2 also repress FT expression. Moreover, TEM1 binds directly to the 5′ untranslated region of FT. A balance between CO and TEM genes has been described as an important mechanism in the transcriptional control of FT particularly in young plants. Other repressors of FT belong to a small group of AP2-like genes, mRNAs of which are targets of miR172. This group includes APETALA2 (AP2), TARGET OF EAT 1 (TOE1), TOE2, SCHNARCHZAPFEN (SNZ) and SCHLAFMÜTZE (SMZ). However, only SMZ is known to bind directly to a region 1.5 kb downstream of the FT stop codon. Fernando Andrés & George Coupland http://www.nature.com/

Structure and function of Florigen and the receptor complex; www.cell.com
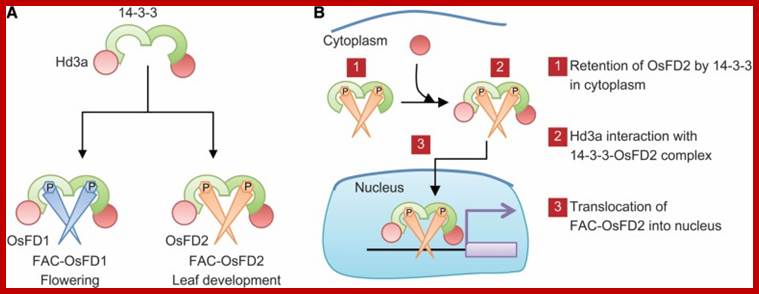
Model of FAC function converted by OsFD1 or OsFD2 (A) and the mechanism of FAC formation including OsFD2 (B). (A) The Hd3a–14-3-3 protein subcomplex serves as the basic component of the FAC. When OsFD1 enters the complex, the resultant FAC-OsFD1 promotes flowering. OsFD2 is the proposed component of FAC to form FAC-OsFD2 that putatively controls leaf development. In this model, the function of FAC can be converted, depending on the function of the OsFDs recruited into the FAC. (B) A part of the OsFD2 protein is localized in the cytoplasm by the interaction with 14-3-3 proteins. When Hd3a interacts with the 14-3-3–OsFD2 complex in the cytoplasm to form a FAC, the complex enters the nucleus to regulate gene expression. Plant cell physiology; In shoot apical cells, conserved cytoplasmic 14-3-3 proteins act as florigen receptors. A hexameric florigen activation complex (FAC) composed of Hd3a, 14-3-3 proteins, and OsFD1, a transcription factor, activates OsMADS15, a rice homolog of Arabidopsis APETALA1, leading to flowering. http://www.ncbi.nlm.nih.gov/

http://www.tanpaku.org/
FD is basic Leucine zipper (bZIP) protein, a transcription factor of Arabidopsis. The c-terminus contains a short motif targeted by Ca2+ dependent protein kinase (CDPKs) of serine/threonine. This phosphorylation is required for interaction of 14-3-3 proteins with FD function. The crystal structure of FAC suggests that FD acts to tether the protein complex on the target promoter element of DNA. FDs from different sources share 14-3-3 protein interaction motifs at their C-terminus, suggests that these FDs are homologous. The ternary complex is known as the Florigin activating complex (FAC) which activates OsMADS15 a MADs domain TF that regulates flowering.
14-3-3 proteins act as intracellular receptors for rice Hd3a florigen; http://www.rcsb.org/

Structure of Florigen Activation Complex Consisting of Rice Florigen Hd3a, 14-3-3 Protein GF14 and Rice FD Homolog OsFD1; http://www.rcsb.org/
Molecular-Genetic Research on Flowering;
With the advent of Arabidopsis as a model plant for molecular-genetic studies, genetic and molecular analyses became popular approaches in studies on flowering. Through mutagenesis, many mutants were isolated in the quantitative long-day plant (LDP) Arabidopsis thaliana. Of interest here are those mutants that exhibit changes in flowering time in comparison with wild-type (WT) plants. Mutants flowering later than WT plants represent a loss-of-function that must involve positive regulators of flowering. Conversely, early-flowering mutants have lost an inhibitor of flowering. These molecular-genetic studies have led to identification of four pathways that regulate flowering in Arabidopsis: the photoperiod, vernalization, autonomous, and GA pathways (e.g., Komeda 2004; Corbesier and Coupland 2005, 2006; Imaizumi and Kay 2006). In the present essay, we will deal mainly with the photoperiod pathway.
The genes called CONSTANS (CO) and FLOWERING LOCUS T (FT) are principlal to long day induced flowering in Arabidopsis. CO encodes a nuclear zinc-finger protein, which in response to LD induces transcription of FT in the phloem of leaves (?).Does it happens in phloem or in leaf tissues, but where in the cells of leafs; is it mesophyll cells, bundle sheath cells or in cells in contact with phloem progenitors. Visualize phloem food conducting vessels are like human lympho system. Neither CO nor FT is expressed in the shoot apex. Expression of CO from a meristem-specific promoter does not enhance flowering, but early flowering is induced in short days (SD) when FT is overexpressed in the shoot apex. Expression of CO from a phloem-specific promoter is sufficient to generate a phloem-mobile stimulus that induces flowering, as shown by grafting experiments between Arabidopsis donor plants over expressing CO and co mutant shoots as receptor (An et al. 2004; Ayre and Turgeon 2004). Because FT must act in the shoot apex in order to elicit flowering, this result gives a strong indication that FT or its product is the signal that moves from an induced leaf to the shoot apex and induces flowering.
FT acts in the shoot apex by forming a complex with the bZIP transcription factor FD. The essential role of FD in flowering is demonstrated by the finding that fd mutants flower late and that FT over expression is partially suppressed by fd (Abe et al. 2005; Wigge et al. 2005). The FT/FD complex activates the downstream genes APETALA1 (AP1) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1); the latter, in turn, activates LEAFY (LFY). So, although FT and FD are produced at different sites, they act together in the shoot apex (Figure). This finding suggests that the FT gene product has to move from the leaf to the shoot apex and strongly implicates the FT gene product as part of florigin, if not florigin itself. The concept of FT as florigin, a protein of 20 Kd how does it induce cells in the SAM. On what cells it acts? Is it through the binding to FT receptors found in the basement cell membranes of SAM or what?

Regulation of Arabidopsis thaliana FT transcription; The genetic basis of flowering responses to seasonal cues; http://www.nature.com/
Transcription of FLOWERING LOCUS T (FT) is a point of convergence of different seasonal cues and is tightly regulated. A 5.7 kb region 5′ of the ATG represents the FT promoter and contains all of the elements that are required to mediate spatial and temporal expression of FT in response to photoperiod. CONSTANS (CO) activates FT transcription in response to long days and was recently demonstrated to bind in vitro and in vivo to the proximal region of the FT promoter. Interestingly, although CO binds to the proximal region of the promoter close to the transcription start site, a more distal region (shown by the dotted arrow in the figure) also seems to be essential for CONSTANS (CO)-dependent FT activation. This upstream region might be recognized by an unknown activator complex that enhances the affinity of CO to bind to the region located in the proximal promoter or is required for CO function. The CCAAT-box-binding complex has been proposed to carry out such a function. GIGANTEA (GI), FLAVIN KELCH F BOX 1 (FKF1) and CYCLIC DOF FACTOR 1 (CDF1), which affect expression of FT by regulating CO mRNA level, are also reported to bind directly to the FT locus. Indeed, tagged CDF1 proteins expressed in the fkf1 mutant strongly associate near the FT transcription start site, suggesting that FKF1 contributes to FT transcriptional activation by removing CDF repressors from its locus. The basic helix–loop–helix transcription factor PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) binds directly to the proximal FT promoter in response to high temperature when it activates FT transcription independently of CO, and another basic helix–loop–helix transcription factor, CIB1, binds to a similar region and activates FT in response to blue light. LIKE HETEROCHROMATIN PROTEIN1 (LHP1) is a direct repressor of FT transcription. In lhp1 mutants, FT mRNA is highly expressed, and flowering is accelerated independently of photoperiod. LHP1 is a chromodomain protein that colocalizes with histone H3 modified by trimethylation on lysine 27 (H3K27me3), which is a repressive chromatin mark formed by Polycomb repressive complex 2 . LHP1 is proposed to be a plant-specific component of Polycomb repressive complex 1, (chromatin immunoprecipitation (ChIP) experiments (such as ChIP followed by microarray (ChIP–chip) and ChIP followed by high-throughput sequencing (ChIP–seq)) showed the pattern of accumulation of these repressive marks along the locus of FT (upper panel of the figure). Both H3K27me3 and LHP1 widely cover the FT locus, conferring its transcriptional repression, whereas those regions that are free of repressive marks have been proposed to constitute a window of open chromatin that is accessible to regulatory factors. Many of the other proteins that bind the FT locus are also transcriptional repressors. The MADS box proteins SHORT VEGETATIVE PHASE (SVP) and FLOWERING LOCUS C(FLC) confer vernalization requirement and repress FT by binding CArG boxes present in its promoter and first intron SVP and FLC are proposed to bind FT in a heteromeric complex. The AP2-like transcription factor TEMPRANILLO (TEM) genes TEM1 and TEM2 also repress FT expression. Moreover, TEM1 binds directly to the 5′ untranslated region of FT. A balance between CO and TEM genes has been described as an important mechanism in the transcriptional control of FT particularly in young plants. Other repressors of FT belong to a small group of AP2-like genes, mRNAs of which are targets of miR. This group includes APETALA2 (AP2), TARGET OF EAT 1 (TOE1),TOE2, SCHNARCHZAPFEN (SNZ) and SCHLAFMÜTZE (SMZ). However, only SMZ is known to bind directly to a region 1.5 kb downstream of the FT stop codon: http://www.nature.com/
Is the Phloem-Mobile Floral Stimulus FT mRNA or FT Protein?
A substance functioning as florigin must fulfill at least the following criteria:
- It must be produced in the leaf, presumably only under inductive conditions for flowering. At least, it would be expected to be more abundant in an induced than in a non-induced leaf.
- It must be phloem-mobile, moving from an induced leaf to the shoot apex.
- It must be required for flowering in all plants.
From the hierarchy of regulatory proteins that controls flowering, it follows that FT is the terminal gene expressed in the leaf, whereas its effect is in the shoot apex (see above; Corbesier and Coupland 2006). Consequently, three possibilities can be envisaged:
- FT mRNA is phloem-mobile and moves from an induced leaf to the shoot apex.
- FT protein moves from an induced leaf to the shoot apex.
- FT in the leaf controls the synthesis of a small compound in the leaf that then moves to the shoot apex, where it induces FT expression to produce FT protein, which complexes with FD. Results relevant to all three possibilities will be discussed below.
Huang et al. (2005) investigated the possibility that FT mRNA is the mobile stimulus. It was shown in Arabidopsis that FT under control of a heat shock promoter was transiently expressed in a single heated leaf and FT mRNA was detected in the shoot apex by RT-PCR 6 hours later. In addition, the heat-treated leaf did induce flowering. Thus, in this work FT mRNA would seem to fulfill some of the criteria of florigen. However, it was later reported that the real-time RT-PCR data were analyzed incorrectly and that in new experiments the movement of FT mRNA from leaf to shoot apex was not detected. Consequently, “…the conclusion that FT mRNA is part of the floral inductive signal moving from leaf to shoot apex” was retracted (Böhlenius et al. 2007). Other authors also failed to obtain evidence favoring FT mRNA as a mobile floral stimulus. In the case of Arabidopsis and rice, FT mRNA as measured by real-time RT-PCR was lower in shoot apices than in leaves (Corbesier et al. 2007; Tamaki et al. 2007). Also, no FT mRNA could be detected in the shoot apex of Arabidopsis by in situ hybridization (Jaeger and Wigge 2007). In Cucurbita, FT mRNA was not detected either in the phloem sap or in the shoot apex (Lin et al. 2007). Finally, in grafting experiments with tomato SFT transcript of the donors did not move across the graft union to the receptor shoots (Lifschitz et al. 2006; Lin et al. 2007).Thus, a rather extensive body of evidence argues against FT mRNA functioning as florigen, although considering the presence of mRNAs in phloem as components of a long-distance signaling network, the possibility cannot be completely ruled out (Lough and Lucas 2006).
Simultaneously with the retraction of the publication on FT mRNA as a phloem-mobile signal, two new publications reported results that FT protein rather than the mRNA is the mobile signal in the phloem that induces floral initiation in Arabidopsis as well as in the SDP rice (Figure 1). To determine the distribution and movement of FT protein, George Coup land’s team at the Max Planck Institute for Plant Breeding Research, fused FT with the gene encoding GREEN FLUORESCENT PROTEIN (GFP) and expressed this construct in ft mutant plants. Expression from phloem-specific promoters demonstrated the presence of FT:GFP not only in leaf phloem, but also in the shoot apex. FT:GFP transgenic plants also flowered earlier than ft plants. Assuming that the promoters employed in these experiments are specific for expression in phloem, these results demonstrate that the FT protein expressed in the leaf phloem moves to the shoot apex to induce floral initiation (Corbesier et al. 2007). FT and FT:GFP were also expressed from the GALACTINOL SYNTHASE (GAS1) promoter, which acts specifically in the phloem companion cells of the minor veins of leaves. In transgenic plants expressing GAS1:FT:GFP, GFP fluorescence was observed only in the minor veins of leaves. Transgenic GAS1:FT plants flowered early, but GAS1:FT:GFP plants flowered as late as ft plants, although the transgene was active in the leaves, as shown by activation of FRUITFULL (FUL). The authors speculated that the fusion protein was not mobile in the minor veins and as a result did not move to the shoot apex and induce flowering. This result confirms that FT protein is the floral stimulus in Arabidopsis and that no secondary product of FT is involved (Corbesier et al. 2007).
In the SDP rice, Heading date 3a (Hd3a) is the ortholog of FT in Arabidopsis. Under SD conditions, Hd3a shows highest expression in leaf blades of rice. Ko Shimamoto’s group at the Nara Institute of Science and Technology, transformed rice with the Hd3a: GFP construct under control of phloem-specific promoters. The transgenic plants flowered early and GFP fluorescence was observed in the vascular tissues of the leaf blade, stem, and shoot apex. Because the Hd3a:GFP construct was expressed only in the phloem of leaf blades, whereas the protein was detected in the shoot apex, it must be concluded that Hd3a protein is moving in the phloem and that Hd3a functions as florigin in rice (Tamaki et al. 2007).
Additional evidence obtained with Arabidopsis further supports the notion that FT protein moves from an induced leaf to the shoot apex. When expressed from a phloem-specific promoter, an epitope-tagged version of FT induced early flowering; the protein was detected by immuno localization in the shoot apex. By contrast, when MycFT was targeted to the nucleus (immobilized), it had no effect on flowering and remained localized in the phloem of the leaf, whereas the constitutive 35S promoter did induce early flowering (Jaeger and Wigge 2007). Furthermore, FT expressed ectopically as a large fusion protein promoted flowering. But in the case of a phloem-specific promoter, flowering was promoted only if the FT protein was released from the complex by a specific protease (Mathieu et al. 2007). Thus, export of FT from companion cells was required and correlated with flowering.
In the experimental approaches used in Arabidopsis to demonstrate the movement of FT protein, FT was expressed as a fusion protein in transgenic plants for ready detection by either confocal microscopy or immunolocalization. It should be realized that expression of FT transgenes is usually much higher than that of the native FT gene. Although all the results are consistent with movement of FT protein from induced leaves to the shoot apex, the presence of native FT protein in the phloem translocation stream was not demonstrated in these experiments. In a close relative of Arabidopsis, Brassica napus, FT protein has been identified in phloem exudate of inflorescence stems (Giavalisco et al. 2006), so that it is reasonable to assume that native FT protein is also present in the phloem sap of Arabidopsis.
Because of its small size, Arabidopsis is not a suitable plant for phloem translocation studies. For this purpose, Cucurbita has been a favorite, especially because phloem exudate can be readily collected from cut stems. However, cucurbits are day-neutral and had not been used for flowering studies until Bill Lucas’s laboratory at the University of California, Davis, surveyed 97 cucurbit accessions. They found one, C. moschata (Cmo), that flowered only under SD conditions. The group tested this species, along with day-neutral C. maxima (Cm), to determine whether long-distance movement of FT is required for flowering. They used the Zucchini yellow mosaic virus (ZYMV) as vector for introducing the FT gene of Arabidopsis (AtFT) into Cmo. Infection with this vector caused flowering of Cmo in LD. The virus was present in developing leaves, but not in apical tissues (Lin et al. 2007). This result demonstrates that the function of AtFT as an inducer of flowering is conserved when expressed in Cmo, and further, that flowering was most likely induced by movement of FT protein from virus-infected leaves to apical regions.
To investigate the role of FT-like (FTL) genes in Cucurbita, two orthologs of FT were isolated from both Cm (Cm-FTL1 and Cm-FTL2) and Cmo (Cmo-FTL1 and Cmo-FTL2). Transcripts of FTL genes were restricted to phloem of stems and leaves, but were not detected in phloem sap. By contrast, the FTL proteins were detected in phloem sap by a combination of liquid chromatography-tandem mass spectrometry. Cmo-FTL2 was approximately 10 times more abundant in phloem exudate than was Cmo-FTL1. Both proteins were present only in phloem sap obtained from SD-grown plants. Although transcript and protein levels of Cmo-FTL2 were much up-regulated in the phloem of stems in SD, an important regulatory mechanism by the photoperiod appeared to be entry of FTL proteins into the phloem translocation stream, in addition to transcriptional control (Lin et al. 2007). Finally, Cmo (receptor) was grafted onto Cm (donor) in long days. All receptor shoots were induced to flower and the CmFTL2 protein was identified in phloem sap collected from Cmo scions. These results show convincingly that FTL proteins are present in the phloem translocation stream of Cucurbita and that they can move across a graft union in a heterograft to induce flowering in a vegetative receptor shoot (Lin et al. 2007). This experiment elegantly demonstrates that transmission of florigen from a donor to a receptor plant is associated with transfer of FT protein from donor to receptor.
FT Protein Is the Universal Signal for Flowering;
The basic tenet of the florigin hypothesis is that florigin is common to all flowering plants. Physiological evidence for the universality of florigin is based on results of grafting experiments between closely related species in which one response type (e.g., a LDP) induces flowering in a related species of another response type (e.g., a SDP). However, this approach has been limited by graft-incompatibility between unrelated species. With the advent of molecular genetics and plant transformation, this barrier can now be readily overcome. Instead of grafting, a specific gene from one species can be “transplanted” into an unrelated species and its role in flowering demonstrated. For example, the SFT gene of day-neutral tomato can substitute for the LD requirement in Arabidopsis (Lifschitz et al. 2006). Likewise, FT expressed in the SDP C. moschata induces flowering under LD conditions (Lin et al. 2007).
One of the criticisms of the universality of florigin was that there were many examples of grafting experiments in which receptor shoots failed to flower (reviewed in Zeevaart 1976). Does this mean non-identity of florigin in the two grafting partners? Work with tomato provides an answer to this question. Transgenic tomato plants over expressing SINGLE-FLOWER TRUSS (SFT), an ortholog of FT, under control of the 35S promoter, were excellent donors, but wild-type tomato could not complement sft mutant plants in grafting experiments (Lifschitz et al. 2006). This result suggests that the failure to induce flowering in receptors is not due to non-identity of florigin, but most likely due to a low level of florigin in the donor and/or rapid decay of florigin in the receptor.

The protein Flowering Locus T and terminal flower TFL; https://blogs.biochem.ncsu.edu

FT from Arabidopsis thaliana; https://cansar.icr.ac.uk; http://swissmodel.expasy.org/
The functions of FT orthologs appear highly conserved in flowering plants regardless of response type, and in monocots as well as in dicots. Although the control of expression of FT varies across response types, the end product, FT protein, appears to be always the same. The evidence obtained so far provides strong support for the universality of FT protein as florigin not only in herbaceous plants, but also in trees (Böhlenius et al. 2006; Hsu et al. 2006). In Lolium temulentum, GAs, specifically GA5 and GA6, have been assigned a role as florigen (Web Essay 25.1). But it is of interest that also in this species LtFT was strongly up-regulated in the leaves after plants had been shifted from SD to LD (King et al. 2006).
The question is often asked: Why did it take so long to elucidate the molecular nature of florigin? There are several aspects to a complete answer. For many years it was assumed that florigin, like the classical plant hormones, would be a small molecule that could be extracted and re-introduced into test plants. Some physiological evidence indicated that in some species florigen has virus-like properties, but techniques to extract nucleic acid or proteins and apply them to assay plants were not available. So, further progress had to await the application of molecular-genetic techniques to studies on physiology of flowering. It then took considerable time before the various flowering pathways had been worked out in Arabidopsis and the pivotal role of FT in flowering became apparent (e.g., An et al. 2004; Ayre and Turgeon 2004; Abe et al. 2005; Wigge et al. 2005; Imaizumi and Kay 2006). Rather than applying extracts to test plants, transgenes could now be expressed and their products, mRNA and protein, could be visualized by techniques of cell biology, or identified by mass spectrometry. It took 70 years, but finally florigin has been identified as FT, a mobile protein of approximately 20 Kda.
Perspective
With the identification of FT protein as florigin, questions regarding its production, transport, and persistence can be studied at the molecular level. For example, is FT permanently activated in plants with localized induction? Does FT induce its own production via a positive feedback loop in species that exhibit the phenomenon of indirect induction of flowering? (see textbook pp. 661–662). These intriguing phenomena, as well as other classical observations on the physiology of flowering, can now be studied from a molecular-genetic perspective. It is expected that results of further studies will provide a solid underpinning for the florigen theory.

Mechanism of flowering by
binding between Florigen and
its receptors in shoot apical
cells
Flowering induction in plants occurs as follows. Florigen synthesized in the leaf is transported through the vascular bundle to the shoot apex, from which flowers come out. In the shoot apical cells, Florigen Hd3a, binds to the 14-3-3 receptor in the cytoplasm. The resulting Hd3a-14-3-3 complex is transported into the nucleus and binds to OsFD1 to form a higher complex, the Florigen activation complex. This complex binds to the promoter of genes that induce the formation of flower buds, and activates the genes that in turn activate a series of genes that promote flowering.

Crystal structure of Florigen activation complex:
The crystal structure of the florigen activation complex, which is a hexamer comprising two each of three proteins, i.e., Hd3a, binds to 14-3-3 to form Hd3-14-3-3-oSfd (acts as florigin activating complex), and OsFD1, and is responsible for the activation of florigen, was determined. DNA-binding sites were simulated by modeling techniques. Red, blue, and green ribbon diagrams represent florigen, the 14-3-3 protein (receptor), and the OsFD1 transcription factor that binds to DNA, respectively. http://www.spring8.or.jp/
Some of the floral induction gene products and their molecular weight:
FT = 20 to 22 kDa,
CO = 46.20 kDa,
SOC = 24kDa,
FD = 31.2kDa,
LHY = 46.58kDa,
AP1 = 255.30.187,
AP2 = 19kDa,
PI = 22kDa,
AGl =27kDa,
FLC = 21kDa,
FLC = 21.5kDa,
FRI = 669 kDa,
Sepallata = 670 kDa,
Agamous like MADs = 95a.a
Apetala1and 2,
Squamosa:
Lipless 1 and 2
PhAP2B, 2C, PhAP2A,
Pistillata,
APA3
ApGLO and ApDEF-21--214a.a
Floral binding protein=FPB
HOX genes
Superman-TF
FRIGIDA
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y, Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056. An, H.L., Roussot, C., Suárez-López, P., Corbesier, L., Vincent, C., Piñeiro, M., Hepworth, S., Mouradov, A., Justin, S., Turnbull, C., and Coupland, G. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626
The Role of Gibberellins: Role of GA in Floral Evocation of the Grass Lolium temulentum; Rod W. King and Lloyd T. Evans, CSIRO, Plant Industry, GPO Box 1600 Canberra, ACT 2601, Australia; September, 2006
Introduction
Almost 70 years ago it was recognized that the leaf is the site of the initial response in daylength-regulated flowering. However, the nature of the factors transported from the leaf to the shoot apex where the flowers form has remained elusive. Here we summarize the evidence from studies with the grass Lolium temulentum, which shows that specific gibberellins (GAs) act as its "floral stimulus." This evidence satisfies five requirements, namely:
The
production of florigenic GAs in the leaf when exposed to a photo inductive long
day
Their
transport from the leaf to the shoot apex
Their
sufficient increase in the shoot apex when floral evocation occurs
Morphological
and molecular changes at the apex, especially involving floral organ identity
genes
Replacement
of the long-day requirement by specific GAs
Our studies over the last 40 years, reviewed by King and Evans (2003), have established a continuous trail of evidence for the gibberellin (GA) class of plant hormone as a floral stimulus in the long-day regulated flowering of the grass, Lolium temulentum. Its long day (LD) photoresponse in the leaf blade involves far-red rich phytochrome-active light, which enhances the expression of a GA biosynthetic enzyme and alters GA precursor and product levels. Most significantly, the florigenic GAs increase in the shoot apex not long after they rise in the leaf, and at the time of the earliest molecular and morphological changes at the shoot apex. Finding the final pieces of this scientific jigsaw has involved GA application studies. Not only was it necessary to show that applied GAs could replace the need for a LD, but by utilizing a range of structurally related synthetic and natural GAs as well as inhibitors of GA synthesis, we have established that some GAs promote inflorescence initiation while others are more active for stem elongation or may participate in the later stages of flowering of L. temulentum.
Gibberellins for Flowering;
In 1956, Anton Lang reported for the first time that applied gibberellic acid (GA3), like vernalization or exposure to long days (LDs), could cause plants of Hyoscyamus niger first to bolt, and subsequently, to flower. His finding was soon confirmed both for other dicot species, for grasses and for GAs other than GA3 (Lang 1957).
Flowering of many temperate grasses is induced by LD or by treatment with GA3. For example, the grass Lolium temulentum requires a single LD for inflorescence initiation, and this requirement can be met in noninductive short days (SD) by a single application of some GAs, either to the leaf blade or to the shoot apex (Evans 1964a, 1969; Evans et al. 1990). However, there are differences between some GAs and LD in this response. Over at least the first three weeks of inflorescence development, there is little or no stem elongation/bolting associated with the LD-induced flowering of L. temulentum and perhaps other grasses—a response duplicated by application of some GAs—but many GAs that induce flowering of grasses also cause early and excessive stem elongation.
The responses to GAs may be similar across dicot and monocot species even though the effects on stem elongation and flowering may occur in a different order, presumably reflecting a different order of gene activation associated with different GAs. For example, GAs that stimulate stem elongation may not be involved in the process of floral evocation in grasses, although they could be involved in subsequent inflorescence development. Conversely, other GAs may be florally effective because they cause inflorescence initiation with little or no effect on stem elongation.
The Search for GAs that are Florigenic but Ineffective for Stem Elongation;
Beginning in the mid 1980s, and in collaboration with Dick Pharis and Lew Mander, we compared the effects of many different gibberellins on floral evocation and stem elongation in L. temulentum. Some, like GA8 and GA9, influence neither process. 16,17-dihydro GA5 promotes flowering while inhibiting stem growth (Evans et al. 1994b), while GA1 and GA3 enhance both and are therefore unlikely to be the floral stimulus in L. temulentum (Evans et al. 1990). However, exogenous GA5 and GA6 are both able to cause floral evocation and induce the early stages of inflorescence development in SD at doses that have no effect on stem elongation (King et al. 1993, 2003). Two questions arise from such findings. Firstly, are either or both GA5 and GA6 agents of floral evocation by one LD in L. temulentum? Secondly, are the more growth-effective GAs such as GA1 and GA4 involved in the subsequent processes of inflorescence development in this grass? To answer these questions, in the following sections we consider evidence based on endogenous gibberellin contents of the leaf and apex of L. temulentum along with information on metabolic pathways involved in GA synthesis and degradation.
Critical Steps in GA Metabolism;
The relevant steps in GA metabolism as summarized by Hedden and Phillips (2000) are shown below:
The key to biological activity among many gibberellins is the closure of the lactone ring with the conversion of GA19 to GA20 and GA24 to GA9. This step involves a 20-oxidase whose activity increases in LD, as shown for spinach and Arabidopsis (Wu et al. 1996; Xu et al. 1997). A multifunctional 3-oxidase completes the conversion of the biologically inactive GA20 or GA9 to bioactive GAs—including GA5 and GA6—and to the 3-hydroxylated GA1, GA3, and GA4. A key inactivation step involves the 2-oxidase responsible for adding a hydroxyl at C-2 to give GA8, GA29, or GA34.
GAs for Floral Evocation: Production in the LD Leaf and Transport to the Shoot Apex;
In L. temulentum, when the leaf is exposed to a single photo inductive LD, expression of the messenger RNA for a 20-oxidase increases dramatically after 16 hours of light and peaks 4 hours later (Figure 2). Most significantly, the timing of the increase in the 20-oxidase in LD matches the minimum duration of light required for floral induction of L. temulentum by one LD. In SD, the expression level of mRNA for this key enzyme is much weaker and peaks 12 hours later during the next 8-hour-daylight period.
|
Expression of a L. temulentum 20-oxidase GA biosynthesis gene. Compared with an 8h short day, LD exposure of the leaf (8h SD extended by 9.5h using incandescent lamps) led to a large increase in gene expression. (Semi-quantitative RT PCR assays from Blundell, MacMillan, and King; unpublished). |
The consequence of increased 20-oxidase activity in L. temulentum leaves is a fall in GA19 levels when the critical day length is reached around midnight, and an equivalent rise in GA20 (GA19 falls from a peak of 35 ng g-1 DW to a minimum of 12 ng g-1 around midnight, while GA20 rises from 3 ng g-1 to 23 ng g-1; King et al. in preparation). GA5, an immediate product of GA20, also increases in LD but not in SD leaves so that the difference by midnight (i.e., after 16 hours of light) is fourfold. GA1 could also be expected to rise in the LD leaves, but this is not apparent until the following day.
The next step, the export of GA5 from the leaf blades and down the leaf sheath has not been directly documented. The difficulty is to isolate specific vascular tissue and to sense potentially small changes in GAs. As an alternative, we have assessed movement from the leaf to the apex of labeled GA5. In LD, 2H4-GA5 applied to the leaf blade was transported intact and quantitatively to the shoot apex (King et al. 2001). Earlier experiments in which the blade and sheath of the single LD leaf were cut off at various positions and times suggest that the LD stimulus is translocated to the shoot apex at a speed of 1–2.4 cm h-1, compared with the simultaneous transport of sugars at about 80–100 cm h-1 (Evans and Wardlaw 1966). Furthermore, evocation can be maximal in shoot apices, which throughout the low intensity day length extension, show no increase in their sucrose content over the 2% found in the vegetative plants exposed to SD (King and Evans 1991). The LD florigenic stimulus in L. temulentum is clearly not sucrose, and an increase in the content of sucrose in the shoot apex is neither necessary nor sufficient for its floral evocation.
Based on its apparent speed of translocation, the LD floral stimulus should begin to arrive at the shoot apex and floral evocation begins on the morning after the LD. Such timing of floral evocation was confirmed by excising shoot apices at various times after the LD, onto media supporting apex development (McDaniel et al. 1991). King et al. (1993) confirmed these results but also found with plants of various ages that shoot apices excised from vegetative plants in SD were induced to flower by supplying GA3 in the medium and could reach a similar stage of flowering to those from plants given one LD but with no GA3 in the medium.
Floral evocation in plants of L. temulentum is associated with an increase on the day after the LD in 32P incorporation into RNA, and of 35S into protein in shoot apices (Rijven and Evans 1967; Evans and Rijven 1967). These increases occur in the dome of the apex and in the sites of future spike lets, down both sides (Knox and Evans 1968). Thus, the timing of these changes fits with the estimated time of arrival of the LD photoperiodic stimulus in the apex (Evans and Wardlaw 1966; McDaniel et al. 1991; King et al. 1993). However, to complete the evidence relating GAs to floral evocation in L. temulentum, it was essential to measure their content in the shoot apex.
The minute size of the shoot apex (<3 µg dry weight) had made it difficult to measure its GA content, but this became feasible through a recent collaboration with Thomas Moritz in Sweden. Using highly sensitive GCMS techniques, he analyzed GAs at subpicogram levels in the shoot apex of L. temulentum. As shown in Table 1, by the end of the daylight period following the overnight LD, the content of the highly florigenic GA5 and GA6 at least doubles in the shoot apex (King et al. 2001, 2003). Moreover, at this time the maximum GA5 concentration reached in the shoot apices is 3 × 10-7M which is close to that needed in an agar medium if shoot apices excised from plants in SD are to flower (about 5 × 10-7M GA5; King et al. 1993). It is highly probable, therefore, that translocation of these two GAs from leaf blades in LD causes floral evocation of L. temulentum.
Changing the Guard: Inflorescence Development and Growth-Active GAs ;
A number of bioactive GAs and precursors including GA1, GA4, and GA9 are notably absent from the shoot apex or minor in SD and on the day after the inductive LD, and remain so for 6–10 days. After exposure of the leaf to 2 or 3 LD, they all increase in content at the apex (King et al. 2001), whereas the content of GA5, GA19, and GA24 falls more rapidly with additional LD. Such additional LDs accelerate inflorescence development (Evans 1958, 1960; Evans and Blundell 1996) and, as well, increase the content of GA1, GA4, and GA9 in the LD leaf (Gocal et al. 1999). Thus, as for floral evocation, for inflorescence development there are increases in GAs in LD but now for a new group of GAs that are highly active for stem elongation.
During inflorescence development, the many fold increase in the shoot apex of the "new guard" GAs—GA1 and GA4—along with evidence that their application can now promote flowering (King et al. 2001), shows their importance for inflorescence development. That a GA input is effective during floral development is also evident from studies with apices induced to flower by one LD, and then excised. Progress to flowering is weak unless GAs are supplied by about six days after the end of the LD (King et al. 1993). Which GAs are important at this time is indicated by the response to application of two growth retardants, Trinexapac Ethyl and LAB 198 999. These retardants block synthesis of GA1 and GA4 by 3-oxidases and inhibit inflorescence development but show no inhibition when applied earlier during floral evocation (Evans et al. 1994a). Thus, whereas GA5 and GA6 are the gibberellins most active in floral evocation of L. temulentum, it is the elongation-active, C-3-hydroxylated GAs such as GA1 and GA4 that play a role in subsequent inflorescence development.
Since GA1 and GA4 can be detected in leaves of L. temulentum—whether from vegetative or floral plants—there must be some mechanism first to exclude them from the shoot apex and then allow them to access the shoot apex late in inflorescence development. As discussed below, this exclusion mechanism may involve: (i) degradation of specific GAs by a 2-oxidase; (ii) localization of a 2-oxidase just below the apex of vegetative plants and; (iii) disappearance of this 2-oxidase during inflorescence development. The implication that some GAs are more readily degraded than others (e.g., GA1 or GA4 vs GA5 or GA6) also provides a focus in the following section for understanding how GA structure affects response.
Structural Considerations: The Lord of the Rings;
The functional groups contributing to GA activity are indicated in Figure below, and from comparisons of many GAs (Evans et al. 1990, 1994 a,b; King et al. 2003; Mander et al. 1998 a,b), it is clear that the structural requirements for florigenicity are quite different from those for stem elongation. As an example, for four GAs—2,2-dimethyl GA4, GA32, GA1, and GA4—although not greatly different in their effectiveness in promoting stem elongation, there is up to a ten thousand fold range in their florigenic activity.
Differences in activity of an applied GA are likely to be determined not only by their ability to resist degradation (see above) but also by structural features altering uptake and transport, their inherent bioactivity, their potential as a biosynthetic substrate, and their capacity to interfere with endogenous GA synthesis and degradation. GA uptake and transport is unlikely to be critical to florigenicity, as GAs could variously promote either or both stem elongation and flowering. Those structural elements favoring florigenicity in particular include:
(i) Formation of the lactone bridge between carbons 4 and 10, so adding a fifth ring to the 4-ringed structure of the biologically inactive GAs. This requirement seems to apply to all the LD grasses whose flowering is induced by gibberellins. For example, GA19 (a 20-carbon precursor GA) is wholly inactive, whereas GA3, GA5, GA7, and several other 19-carbon GAs have reported florigenic activity in other species besides L. temulentum. However, formation of the lactone ring itself is insufficient, and a further step involving activity of a 3-oxidase enzyme is also essential (see iii).
(ii) A free carbon-7 carboxy group is an absolute requirement for all plant GA responses and may reflect ability to bind to a GA receptor.
(iii) Hydroxylation at carbons 3, 12, 13, and 15 and/or the presence of a C-1, 2 double (C-C) bond (as in GA3), or a 2,3 double bond (as in GA5), or a 2,3 epoxide (as in GA6).
(iv) Absence of a C-2 hydroxyl.
While features listed under (iii) may enhance the stability of a GA against inactivation by 2β-hydroxylation, structural elements at C-2 are the most significant in this regard. A good comparison involves the responses to applications of three closely related GAs: the highly florigenic and growth active 2,2-dimethyl GA4, the modestly florigenic but reasonably growth-active 2α-methyl GA4, and the nonflorigenic, growth-active GA4 (Evans et al. 1990). Because it lacks any C-2 structural elements, 2-hydroxylation of GA4 is highly likely and this apparently rings the death knell for any florigenic activity. GA4 is taken up by—and transported in—the plant as it is highly active for stem elongation but quite clearly it is not transported intact into the shoot apex. The most likely explanation, for this observation would involve the presence of a concentrated zone of 2-oxidase just below the vegetative apex, and while this has yet to be demonstrated for L. temulentum, such a highly localized zone of 2-oxidase RNA expression has been reported recently for rice (Sakamoto et al. 2001).
The concept for rice of the exclusion of 2-hydroxylation-sensitive GAs from the shoot apex fits well with the structures of the GAs that are naturally found in the shoot apex of L. temulentum. Neither of the readily 2-hydroxylated GAs we examined, GA1 and GA4, was detectable in the shoot apex of vegetative plants or for about the first week after floral evocation (King et al. 2001) although their content increased in LD leaves (Gocal et al. 1999). By contrast, we detected highly florigenic but weakly growth-active GAs—such as GA5 and GA6—these GAs probably being protected from C-2 hydroxylation by the C-2,3 double bond in GA5 and the C-2,3 epoxide in GA6. High florigenicity with limited growth activity for GA32 also fits with its having a C-1,2 double bond. Later, as floral development proceeds GA5 and GA6 decrease in the apex while there is a dramatic increase in GA1 and GA4 (King et al. 2001) and it is at this stage of flower development that the localized band of 2-oxidase disappears from just below the rice inflorescence (Sakamoto et al. 2001).
Apparently, the localized band of 2-oxidase activity protects the shoot apex from "growth-active" GAs, so guaranteeing the integrity of the apex during vegetative growth, floral evocation, and early floral differentiation. Later, during inflorescence development, disappearance of the 2-oxidase barrier allows the influx of highly growth-active GAs.
Explaining a Paradox: Flowering Promotion by GA Biosynthesis Inhibitors;
Classic studies by Baldev and Lang (1965) with the rosette LDP Samolus parviflorus showed that flowering in LD is inhibited when GA synthesis is blocked using either of the GA biosynthesis inhibitors, Amo-1618 and Cycocel (CCC). Such results supported a role for GAs in the LD response. Paradoxically, however, with L. temulentum, several "anti-gibberellins"—which might be expected to inhibit floral induction—actually promote it, and act synergistically with some GAs. For example, CCC alone did not change the flowering response to one LD, but greatly enhanced the promotive effect of GA3, although other "anti-gibberellins" such as B9 and Amo 1618 gave the expected inhibiting effect on flowering (Evans 1969). With the acylcyclohexanedione inhibitors, LAB 198 999 and Trinexapac Ethyl, promotion of flowering could reflect an interference with the 2-oxidase. Whether or not this enzyme is localized below the vegetative shoot apex, endogenous or applied bioactive GAs would be spared from inactivation. Another explanation involves either or both inhibition of the 3-oxidase(s) responsible for conversion of GA20 to GA1 and inhibition of the 2-oxidase involved in converting GA20 to inactive GA29. By inhibiting the conversion of GA20 to GA1 or GA29, LAB 198 999 and Trinexapac Ethyl treatments could promote GA20 conversion to GA5, thereby accounting for their strongly promotive effects on floral evocation in L. temulentum. Their subsequent inhibitory effects on inflorescence development (Evans et al 1994a) when synthesis of GA1 and GA4 becomes important and it fits with their known action as 3-oxidase competitors. The greater inhibition of the 2-oxidase by the derivative 16,17-dihydro GA5 than by LAB 198 999 (Junttila et al. 1997), indicates further interesting effects of "ant gibberellins," which preclude drawing too simple an interpretation of how they may alter GA metabolism.
The Genes Involved at the Shoot Apex ;
Of the various genes known to be involved in floral determination and differentiation, the earliest to be expressed in the shoot apex of L. temulentum is LtCDC2, which greatly increases in activity in the future spikelet sites by the afternoon after the long day, just when floral evocation is completed (Gocal 1997). By the next day, the API-like gene LtMADS2 and a related gene LtMADS1 have also increased their expression at the spikelet sites and subsequently at the floret sites (Gocal et al. 2001).
Whereas the LEAFY (LFY) gene is expressed early in the floral apices of Arabidopsis, Gocal et al. (2001) found it not to be expressed until about 12 days after LD induction in L. temulentum—possibly associated with the great difference between the two species in the timing of stem elongation vis-à-vis floral evocation, and, as well, in the GAs involved.
Conclusion
For no other species is there such a complete and consistent trail of evidence on the identity of the LD floral stimulus as in L. temulentum, from the day length-sensitive, physiologically mobile GA5, and probably GA6, in the leaf blades, translocated intact and at appropriate velocity to reach the shoot apex in sufficient concentration to effect floral evocation at the clearly identified time.
Moreover, several previously puzzling features of this investigation, especially those relating to the great range in florigenicity among the gibberellins and to the synergistic effects of several inhibitors of GA synthesis, can now be resolved. If, as in rice, the vegetative shoot apex in L. temulentum is protected by a ring of 2-oxidase, a very coherent picture of the control of flowering in L. temulentum by gibberellins emerges. In addition, such localized action of the 2-oxidase explains much of the variation among bioactive GAs in their relative promotion of floral evocation vis-à-vis stem elongation.
Our findings also highlight a feature of the growth habit of temperate grasses, which may have been crucial to their evolutionary success when close-grazed by ungulates. Except for the relatively brief stage when the inflorescences must grow upwards for wind pollination and seed dispersal, the terminal meristems are kept close to the ground by the absence of stem elongation until the later stages of inflorescence development. The initial involvement at floral evocation of GAs such as GA5 and GA6, which do not cause stem elongation at concentrations that are florigenic, and the initial exclusion from the apex of GAs such as GA1 and GA4, which do cause stem elongation, presumably aids survival under both close-grazing and adverse environmental conditions. Such an evolutionary explanation is required, because stem elongation per se is not antagonistic to flowering. Applied 2,2-dimethyl GA4, for example, not only induces flowering but also causes massive stem elongation.
For a number of reasons, our findings that specific GAs are florigens in grasses, cannot be generalized to all species. Based on their evolutionary relatedness (see Kellogg 2001), we believe our findings are applicable to other temperate grasses and cereals, which retain a LD response—as most do (Evans 1964b; Heide 1994). However, the warm climate grasses and cereals—including rice, corn, and sugarcane—often have no photoperiodic response and if they are sensitive, they often respond to SD, which may lead to decreases, not increases, in GA content. Where a species is insensitive to photoperiod, a role for GAs appears unlikely and one of many other florigenic factors must be limiting. In temperate, LD-responsive dicots, GA increases have often been documented but it is also clear that their primary action may be on stem elongation rather than on flowering. This latter observation is especially interesting as the GAs detected in dicots have always been the growth-active ones, and the florigenic but weakly growth-active GAs have yet to be examined in such dicots. Nevertheless, there is no reason at present to suggest that the growth-inactive GAs that are florigens in grasses play any role in flowering of dicots. Baldev, B., and Lang, A. (1965); A mer. J. Bot. 52: 408–417.
References
and Evans, L. T 1958,1960 and 1964a; Control of flower formation by growth retardants and gibberellins in Samolus parviflora, a long-day plant. A mer. J. Bot. 52: 408–417.
Signals produced in leaves are transported to the shoot apex where they cause flowering. Protein of the gene FLOWERING LOCUS T (FT) is probably a long day (LD) signal in Arabidopsis. In the companion paper, rapid LD increases in FT expression associated with flowering driven photosynthetically in red light were documented. In a far red (FR)-rich LD, along with FT there was a potential role for gibberellin (GA). Here, with the GA biosynthesis dwarf mutant ga1-3, GA4-treated plants flowered after 26 d in short days (SD) but untreated plants were still vegetative after 6 months. Not only was FT expression low in SD but applied GA bypassed some of the block to flowering in ft-1. On transfer to LD, ga1-3 only flowered when treated simultaneously with GA, and FT expression increased rapidly (<19.5 h) and dramatically (15-fold). In contrast, in the wild type in LD there was little requirement for GA for FT increase and flowering so its endogenous GA content was near to saturating. Despite this permissive role for endogenous GA in Columbia, RNA interference (RNAi) silencing of the GA biosynthesis gene, GA 20-OXIDASE2, revealed an additional, direct role for GA in LD. Flowering took twice as long after silencing the LD-regulated gene, GA 20-OXIDASE2. Such independent LD input by FT and GA reflects their non-sympatric expression (FT in the leaf blade and GA 20-OXIDASE2 in the petiole). Overall, FT acts as the main LD floral signal in Columbia and GA acts on flowering both via and independently of FT.
Summary of findings here and in the companion paper of positive effects (arrows) on flowering and CO/FT for two commonly used LD photoresponses. This schematic incorporates effects on FT and flowering of: mutants; gene silencing; change in light intensity; and a block to photosynthesis. Predominantly, in LD, photosynthetic sucrose amplifies CO/FT expression (see companion paper) while phytochrome acts directly and also via GA, which plays a permissive and, often, non-limiting role. There is also a direct but lesser LD-mediated increase in GA supply via the petiole response to FR-rich light. A dashed arrow indicates a potential step of regulation, and weaker responses are indicated by thinner arrows. The electronics symbol for a speaker is used to show sucrose amplification of CO/FT expression; http://jxb.oxfordjournals.org/
GA4 applied to ga1-3 shows an FT-independent effect on flowering in SD and a permissive effect involving FT expression in LD. A 10 μl drop of GA4 [1 mM in 20% ethanol: water (v:v)] was applied to each of three leaves on consecutive days either in SD or at the start of a far-red-rich LD (LD-FR). Plants of ga1-3 flowered, bolted, and leaves grew (A). Its FT expression increased most after GA treatment in LD (B), and (C) shows the effect of GA4 on FT expression in Columbia. Prior to treatment, the plants of ga1-3 had been grown in SD for 12 weeks and those of Columbia for 5 weeks. The low intensity FR-rich LD exposure was for 2 d. GA4 was applied 8 h after starting the day, and leaf blades were harvested 19.5 h later for assays of FT expression (leaves harvested at 16 h showed similar increases; not shown). There was no effect of solvent application on flowering or gene expression (not shown). All FT expression was normalized to the value in SD without GA application. The means and SE were based on three replicates for FT assays and 10 replicates for flowering time.
Intergrative pathway;
In Arabidopsis, the four floral pathways converge through genes of the integrative pathway. The activation of floral integrators, such as AtFLOWERING LOCUS T (AtFT) and AtSUPPRESSOR OF CONSTANS 1 (AtSOC1), in turn, lead to the activation of floral meristem identity genes such as LEAFY (LFY) and APETALA1 (AP1). We did not observe differential expression of TaFT (see above discussion of TaVRN3). On the other hand, transcripts for wheat genes that share sequence similarity with AtSOC1 did show differential expression. Zhao et al. identified seven MADS-box genes (TaAGL1, TaAGL7, TaAGL18, TaAGL20, TaAGL21, TaAGL23 and TaAGL38) that, upon phylogenetic analysis, were placed in the SOC1-like clade of MADS-box genes. Only three probe-sets on the wheat array corresponded with these seven genes (see Additional file 5). The gene TaAGL7 corresponds with the probe set Ta.25343. The genes TaAGL1, TaAGL18 and TaAGL23 (the most similar to AtSOC1) all correspond to one probe-set (Ta.21250), and TaAGL20, TaAGL21 and TaAGL38 (the least similar to AtSOC1) to another (TaAffx.19661) – given the high sequence similarity between the genes in the respective groups, they are probably homoeologues and so we cannot report on their individual behavior. However, the three probe-sets that correspond to the genes of the SOC1 clade of MADS-box genes all evidenced essentially the same profile of abundance (data not shown). That is, in both leaf and crown tissue of all three varieties, transcript increased slightly.
Photoperiod pathway;
The principal components of the photoperiod pathway are conserved in the monocots and, more pertinent to this discussion, in the cereals On the Wheat Genome Array, there are probe-sets that correspond to many of the genes belonging to the photoperiod pathway. In Arabidopsis, AtCONSTANS (AtCO) encodes a transcription factor that activates genes required for floral initiation. It integrates circadian clock and day-length signals and, under long-days, activates the floral promoters AtFT, AtSOC1 and AtLFY [8]. The two circadian clock genes AtLHY and AtTOC1 influence the expression of AtCO. They form part of a feedback mechanism, each directly affecting the expression of the other: AtLhy.p is a repressor of AtTOC1, and AtToc1.p is required for the expression of AtLHY [8]. The cyclic expression of these two genes, which occurs over a 24 hour period, entrains that of AtGIGANTEA (AtGI). This latter activates AtCONSTANS.
In this study, the profiles of abundance for TaLHY and TaTOC1 were complementary to each other, as one might expect from their relationship to each other in circadian cycling. In crown tissue, transcript of TaLHY increased in abundance and then declined; conversely, that of TaTOC1 declined and later increased. The profile for TaGI was very similar to that of TaTOC1 (see Additional file 4). Given that in both rice and Arabidopsis, GI is a promoter of CO expression [8], one might have expected the transcript for HEADING DATE 1 (TaHD1), the supposed wheat orthologue of AtCONSTANS, to follow the profile of TaGI. However, it did not show differential expression in this study. Interestingly, other transcripts that appear to be members of the CONSTANS-like family of genes exhibited profiles that did reflect those of TaLHY, and TaTOC1, and TaGI. In particular, a sequence highly similar to barley CONSTANS-like 9 (the most divergent of the barley CONSTANS-like genes which has no counterpart in Arabidopsis [35]) had a profile of abundance very similar to that of TaLHY (see Additional file 4). A transcript with similarity to CONSTANS-like 1 in Lolium perenne, a gene which has been reported to increase after extended periods of exposure to cold [36], had a profile of transcript abundance that echoed that of TaTOC1 and TaGI. Ciannamea et al., using a similar experimental approach to that used in this study, suggested that the profile of transcript abundance for LpCOL1 was suggestive of the gene being involved in the vernalization response [36]. We observed a very similar profile of responses in both the cold treated plants and the controls. This would suggest that this gene in wheat is responding to shortening day length.
GA pathway;
The Affymetrix array doesn't include probe-sets for AtGA1 (codes for ent-copalyl diphosphate synthase) or AtGA INSENSITIVE (the wheat orthologue is REDUCED HEIGHT B1 [RHT B1]). There is a probe-set for AtRGA1 (the wheat orthologue of RHT D1), but we did not observe differential accumulation of this gene. However, there was a clear genotype-dependent, cold response of some components of the gibberellin pathway: transcripts for ent-kaurene synthase and ent-kaurene oxidase (correspond to AtGA2 and AtGA3, respectively), showed leaf specific accumulation (> 20-fold increase after 12 weeks) in the two winter varieties and no response at all in Paragon (Figure 4). This result was confirmed by qRT-PCR (Pearson correlation = 0.99). Ent-kaurene synthase and ent-kaurene oxidase are two of the principal enzymes of the gibberellin biosynthetic pathway [30,31]. Thus, given the profiles of abundance that we observed, one might assume that, in the two winter varieties, there was an increase in gibberellins. In Arabidopsis, gibberellic acid (GA) activates the expression of AtSOC1(SUPPRESSOR OF OVER EXPRESSION OF CO 1) [32], an important integrator of several flowering pathways (discussed below), which in turn promotes flowering through its action on floral meristem identity genes or their products (Komeda 2004). What's more, Moon et al. [32] report that the gibberellin pathway is the only pathway to promote flowering under short days. Thus, it would appear that we have evidence to show that in wheat the gibberellin pathway functions in a similar manner to that in Arabidopsis, and that as a consequence of vernalization under short-days it tends to promote flowering. However, the complete lack of response in the spring variety, Paragon, is intriguing: does the gibberellin pathway not function to promote flowering in spring varieties of wheat.
Vernalization pathway:
Prolonged exposure to low temperatures (vernalization) accelerates the transition to reproductive growth in many plant species, including the model plant Arabidopsis thaliana and the economically important cereal crops, wheat and barley. Vernalization-induced flowering is an epigenetic phenomenon. In Arabidopsis, stable down-regulation of FLOWERING LOCUS C (FLC) by vernalization is associated with changes in histone modifications at FLC chromatin. In cereals, the vernalization response is mediated by stable induction of the floral promoter VERNALIZATION1 (VRN1), which initiates reproductive development at the shoot apex. We show that in barley (Hordeum vulgare), repression of HvVRN1 before vernalization is associated with high levels of histone 3 lysine 27 trimethylation (H3K27me3) at HvVRN1 chromatin. Vernalization caused increased levels of histone 3 lysine 4 trimethylation (H3K4me3) and a loss of H3K27me3 at HvVRN1, suggesting that vernalization promotes an active chromatin state at VRN1. Levels of these histone modifications at 2 other flowering-time genes, VERNALIZATION2 and FLOWERING LOCUS T, were not altered by vernalization. Our study suggests that maintenance of an active chromatin state at VRN1 is likely to be the basis for epigenetic memory of vernalization in cereals. Thus, regulation of chromatin state is a feature of epigenetic memory of vernalization in Arabidopsis and the cereals; however, whereas vernalization-induced flowering in Arabidopsis is mediated by epigenetic regulation of the floral repressor FLC, this phenomenon in cereals is mediated by epigenetic regulation of the floral activator, VRN1.
Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene (W. James Peacock, April 1, 2009). Plants respond to seasonal cues, such as temperature and day-length, to ensure that flowering coincides with favorable conditions. Prolonged exposure to low winter temperatures (vernalization) accelerates the progression from vegetative to reproductive growth in many plant species, including the temperate cereals (such as wheat and barley) and dicot species (such as Arabidopsis) (1–3). In both these lineages, plants retain a “memory” of the prolonged cold of winter, which stimulates flowering when days lengthen during spring (1–3). The memory of cold is then reset in the next sexual generation to ensure progeny are competent to respond to vernalization (1–3).
In Arabidopsis, the vernalization response is mediated by epigenetic regulation of the floral repressor, FLOWERING LOCUS C (FLC), which encodes a MADS-box transcription factor that represses genes involved in floral initiation, including SUPPRESSOR OF CONSTANS 1 and FLOWERING LOCUS T (FT) (1, 4–6). FLC is expressed before vernalization and delays flowering, but its expression is repressed by vernalization (1, 4). FLC remains repressed when plants are subsequently exposed to warm temperatures, allowing activation of FT, which promotes flowering (1, 4). The stable down-regulation of FLC by vernalization is associated with an increase in the levels of repressive histone modifications at FLC chromatin, such as histone H3 lysine 27 di- and trimethylation (H3K27me2, H3K27me3), histone H3 lysine 9 dimethylation, and histone H4 arginine 3 symmetrical dimethylation, as well as the loss of histone modifications associated with active transcription, such as histone H3 acetylation and histone H3 lysine 4 di- and trimethylation (H3K4me2, H3K4me3) (7–13). Repression of FLC by vernalization involves the vernalization-dependent association of Polycomb-Group (PcG) complexes to FLC chromatin, which are required for addition and maintenance of H3K27me3 at FLC (14, 15). Taken together, these studies indicate that vernalization induces an alteration of FLC chromatin state from actively transcribed to stably repressed (7–15). The cellular memory of transcriptional repression of FLC is maintained during successive cell divisions by mitotic inheritance of repressive histone modifications at the gene (11), but active FLC transcription is restored in progeny, ensuring that the next generation is competent to respond to vernalization (1, 4, 16).
In temperate cereals, the vernalization response is mediated by the stable induction of a floral promoter, VERNALIZATION1 (VRN1) (3, 17–19). VRN1 encodes a FRUITFULL-like MADS-box transcription factor required for the initiation of reproductive development at the shoot apex (20–22). In vernalization-requiring cereal plants, VRN1 is expressed at low levels and is induced by vernalization, with the level of expression being dependent on the length of cold exposure (17–19, 23–25). VRN1 expression remains high when plants are exposed to warm temperatures following vernalization, and promotes the transition to reproductive development (17–19, 23–25). VRN1 down-regulates the floral repressor VERNALIZATION2 (VRN2), and allows long-day induction of the floral activator FT to accelerate subsequent stages of floral development (3, 24–26).
The vernalization response of VRN1 shows characteristics of epigenetic regulation, in that VRN1 is induced by vernalization, expression is maintained following vernalization, and the prevernalization level of VRN1 expression is reset in the next generation (17–19, 23–25). In this article we analyze the effect of vernalization on the levels of histone modifications at the barley (Hordeum vulgare) VRN1 gene (HvVRN1). Our study indicates that vernalization-induced flowering in cereals is mediated by epigenetic regulation of VRN1 chromatin state. Our results suggest that regulation of the histone methylation status of VRN1 chromatin is important for repression of VRN1 before vernalization, for activation of VRN1 by vernalization, and for maintaining a memory of vernalization following cold exposure
Environmental signals that favor GA synthesis leads to the bonding of GA to its receptor and interact with their binding proteins, where the regulator of GA called RGA gets degraded through proteosomes. Now the GA signals SOC1/PFP/GAMYB like components that lead to the production FT and FT with FD acts on apical meristem
This figure shows how the GA synthesis leads activation of different response factors that leads to the synthesis of SOC1 and FT, where FT acts on AP1 (?) and SOC1 acts on LFY (?) which ultimately act on flower identity genes.
Below some general and specific flowering pathways have been given and they are self illustrative.

The role of GA in stem elongation seed germination and flower development. www.5e.plantphys.net

The effect of combinatorial expression of gene in the presence and absence of VRN on phenotype is wells illustrated. The figure below shows even Rice palnts produce a FT homolog called Hd3a and it has the same effect as FT of dicots. Dev.biologist.org.

'Quartet Model’, MADS box proteins role in floral development; According to this model, the identity of the different floral organs — sepals, petals, stamens and carpels — is determined by four combinations of floral homeotic proteins known as MADS-box proteins1, 5, 8. The protein quartets, which are transcription factors, may operate by binding to the promoter regions of target genes, which they activate or repress as appropriate for the development of the different floral organs. According to the model, two dimers of each tetramer recognize two different DNA sites (termed CArG-boxes, shown here in grey) on the same strand of DNA, which are brought into close proximity by DNA bending. The exact structures of the multimeric complexes of MADS-box proteins controlling the identity of flower organs are still hypothetical; question marks denote components whose identity is especially uncertain. Proteins: AG, AGAMOUS; AP1, APETALA1; AP3, APETALA3; PI, PISTILLATA; SEP, SEPALLATA.www.nature.com FT homolog Hd3a operates in rice plants, they move from leaves to stem apex, Hd3a in combination with FD (?) induce flowering; www.pcb.org