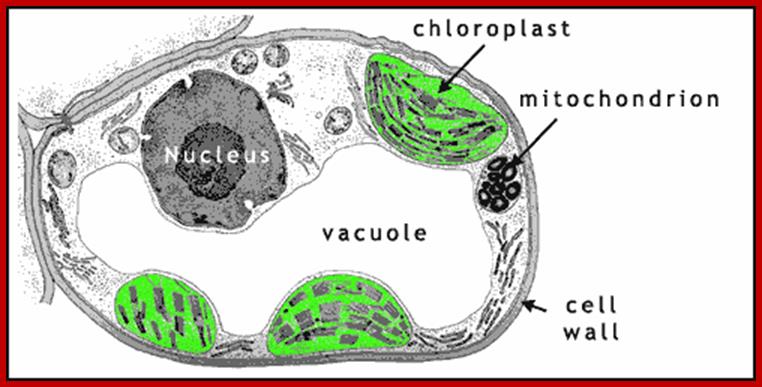
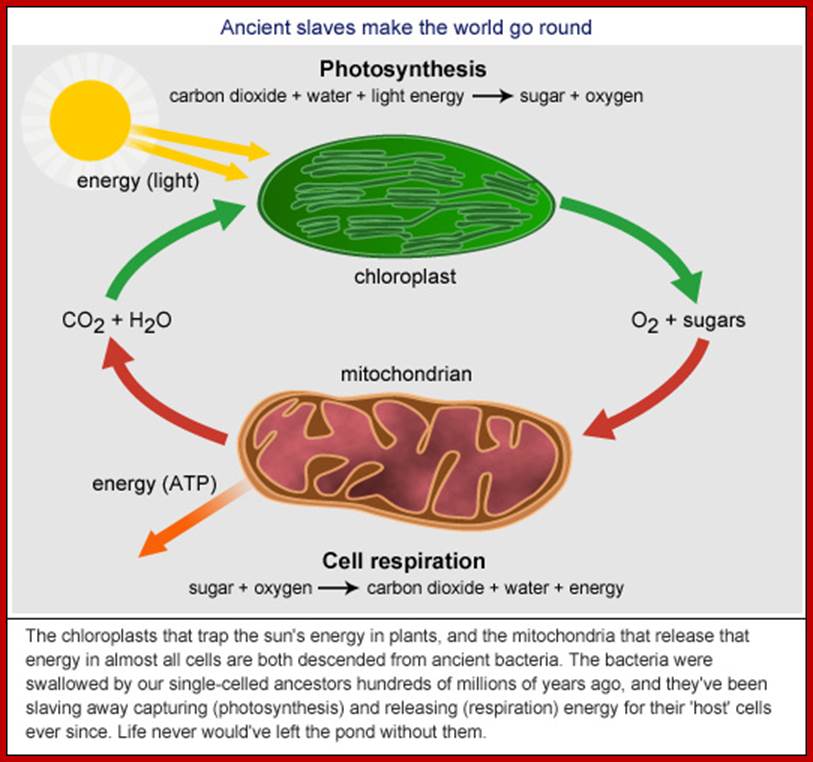
Mitochondria
Mitochondria were first observed by Altman (1894) as filamentous structures and he called them as bioblasts, but Brenda (1897) called them as mitochondria. Unlike chloroplasts they are found in almost all eukaryotic organisms. They are very important energy rich ATP producing organelles and make it available for other biological processes in the form of energy rich compounds like ATP and NADH2 and NADPH2. It also directs fatty acid oxidation products for ATP synthesis. As an endosymbiont mitochondria originated from bacterial cells approximately 1.2 to 2 billion years ago. Origin by a process called symbiogenesis articulated by a Russian Botanist Konstantin Mereschowsky in 1910. A recent study by researchers of the University of Hawaii at Manoa and the Oregon State University indicates that the SAR11 clade of bacteria shares a relatively recent common ancestor with the mitochondria existing in most eukaryotic cells. Cyanobacteria and α-proteobacteria are the most closely related free-living organisms to plastids and mitochondria respectively. All eukaryote contain mitochondria in their cells, except red blood cell. With endosymbiosis the bacterial cells lost their cell wall, but added one more membrane and lost most of its DNA to host eukaryotic cell, but remained together cooperatively. At least in human population both males and females have mitochondria, but inherited to offspring’s only from females.
http://biogenevent.weebly.com /http://bio100.class.uic.edu/ http://learn.genetics.utah.edu/content/begin/cells/organelles/
https://universe-ureview.caUniverse https://
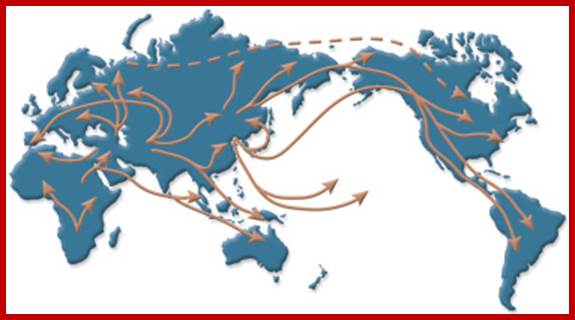
Analysis of mitochondrial DNA from people around the world has revealed many clues about ancient human migration patterns. http://learn.genetics.utah.edu/
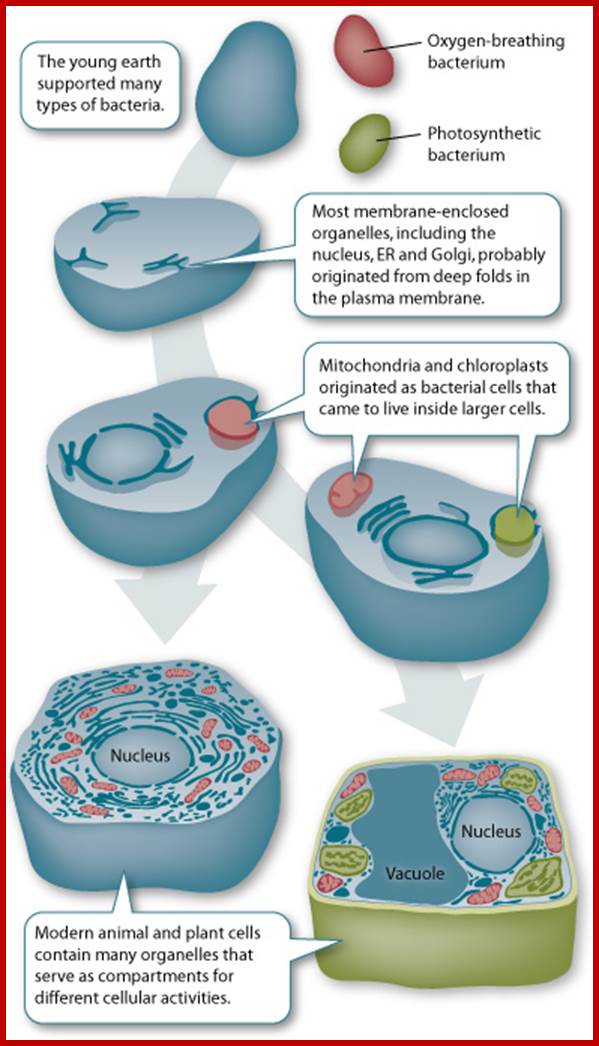
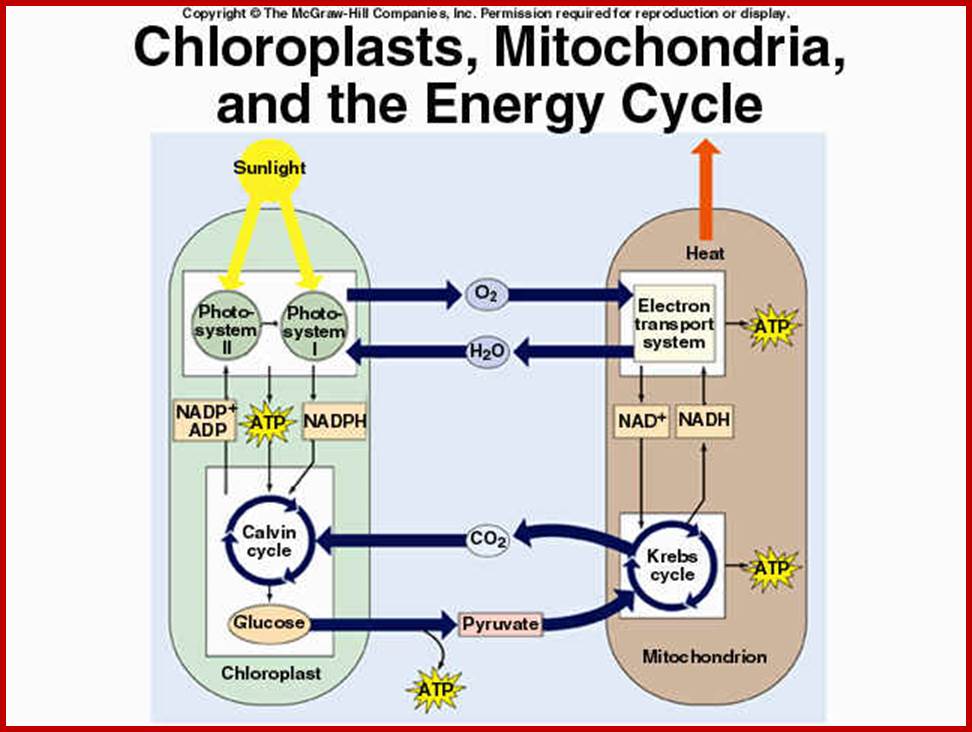
Schematic representation, as shown in the above diagrams, of endosymbiosis, called ‘symbiosgenesis’ leading to the appearance of mitochondria; then there is succession of endosymbiosis with cyanobacteria, what will lead to the creation of a second new organelle specific for plant cell; Chloroplast.; the uniqueness of both mitochondria and chloroplast is that they replicate independently from nuclear DNA replication- for they have their own DNA and their own mechanism of replication, however they require nuclear inputs also for in the past when they are taken in they have lost most of their DNA to the nucleus, but they have coordination with each other. It is estimated that primary plastid s in eukaryotes have believed to have occurred approximately 1.5 billion years ago. It is controversial indeed. Chloroplast endosymbiosis occurred after mitochondria.
The idea of endosymbiosis was first proposed by Konstantin Mereschkowski, a prominent Russian biologist, in 1905. He coined the term "symbiogenesis" when he observed the symbiotic relationship between fungi and algae.
The term "endosymbiosis" has a Greek origin (endo, meaning "within"; syn, meaning "with"; and biosis, meaning "living"), and it refers to the phenomenon of an organism living within another organism. In 1923, American biologist Ivan Wallin expanded on this theory when he explained the origin of mitochondria in eukaryotes (Wallin 1923). However, not until the 1960s did Lynn Margulis, as a young faculty member at Boston University, substantiate the endosymbiotic hypothesis. Based on cytological, biochemical, and paleontological evidence, she proposed that endosymbiosis was the means by which mitochondria and plastids originated in eukaryotes (Sagan 1967; Margulis 1970). In those days, the research community viewed her unconventional idea with much skepticism, but her work was eventually published in 1967 (Sagan 1967) after being rejected by fifteen scientific journals! Today, endosymbiosis is a widely accepted hypothesis to explain the origin of intracellular organelles. http://www.nature.com/
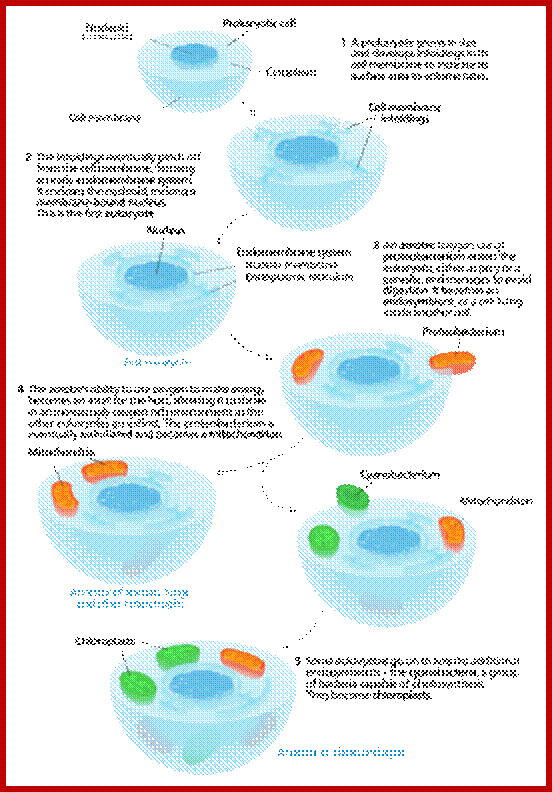
One model for the origin of mitochondria and plastids. https://en.wikipedia.org/

http://www.enchantedlearning.com/galleryhip.comwww.cas.miamioh.edu
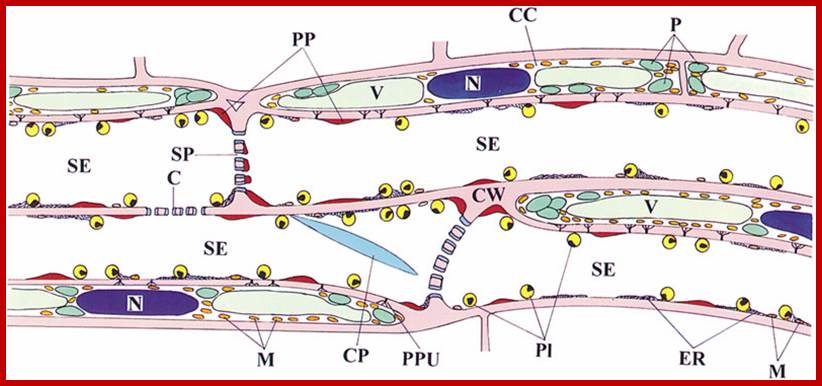
Mitochondria in Sieve cells (M) of Vicia faba; sieve tube cell cross wall is perforated through which Plasmodesmata pass though which provides symplastic connection;. One finds protoplasmic layers next to Plamamembrane, companion cells are adjacent and tightly adhered and one finds a symplastic and as well as aplasic liquid movement, also one finds protein bodies called Pi, endoplasmic reticulum is visible and even mitochondria are found; plastids are also found, Apparently no nucleus is found. In some case certain sieve tube cell associated parenchyma cells are transformed into Transfer cells; http://public.wsu.edu/
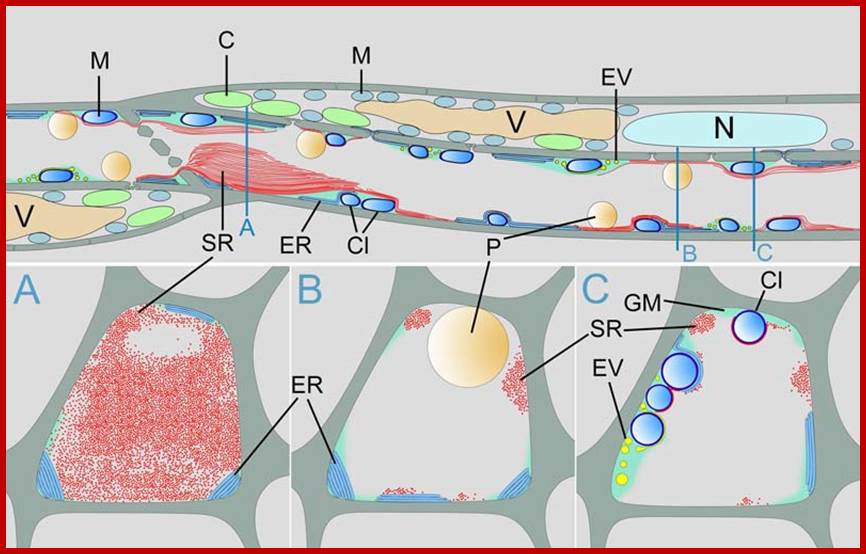
Schematic drawing of sieve tube structure in Arabidopsis thaliana; C = chloroplast, Cl = clamp proteins, ER = endoplasmic reticulum, EV = electron dense vesicles, GM = ground matrix, M = mitochondrium, N = nucleus, P = plastid, SR = SEOR1 filaments, V = vacuole. From Froelich et al. 2011. Phloem ultrastructure and pressure flow: SEOR protein agglomerations do not affect translocation. Plant Cell . Plant Cell doi/10.1105/tpc.111.093179 Copyright American Society of Plant Biologists; http://public.wsu.edu/
https://mortada8.wordpress.com/2009
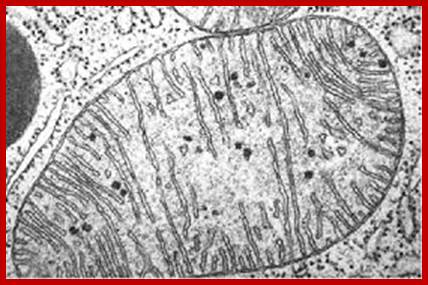
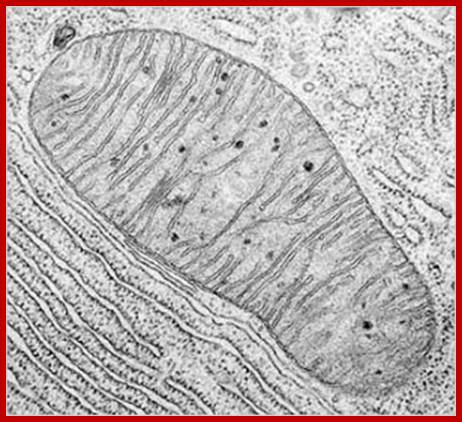
Mitochondria are the cells’ microscopic power plants, producing ATP which powers your muscles; https://roboplant.wordpress.com
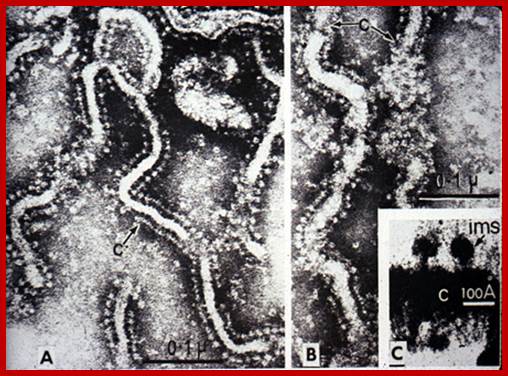
EM of mitochondria- showing inner membrane bond Elementary particles involved in ATP production; http://lifesci.dls.rutgers.edu/

Elementary particles (ATP synthases); http://biology4isc.weebly.com/
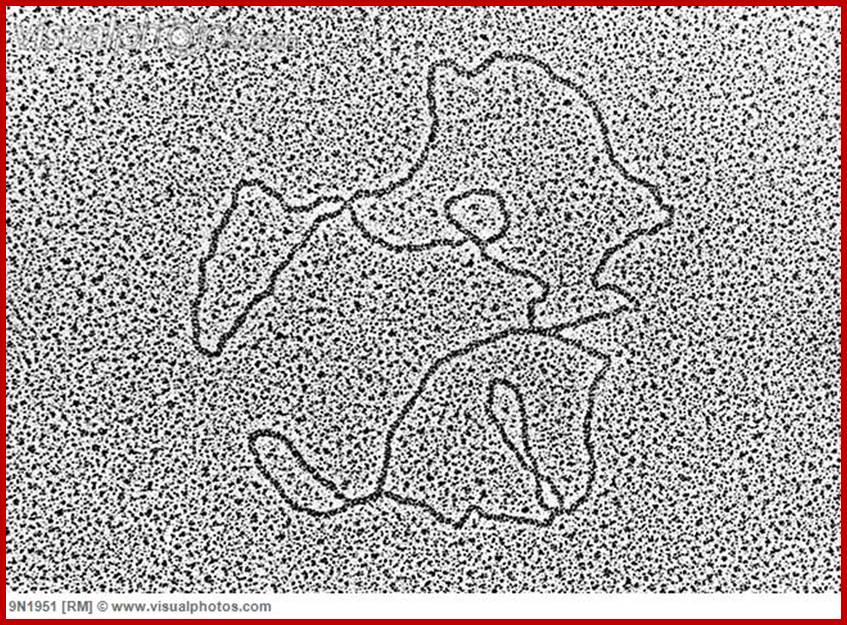
Mitochondrial DNA circular; www.visualphotos.com
Shape sand Size:
Using specific stains or fluorescent dyes, mitochondia can be observed cytologically. In the presence of janus green they look like small rod shaped, greenish blue structures. The shape of mitochondria is never constant; at one moment they look like small bacterial shaped or tubular shaped structures of 0-.5 mm to 7 mm and in next few moments, they appear as long filaments or vesicles. A movie on mitochondria, pictured to show its variability in structure with time, it is amazing to observe how the mitochondria divide and fuse with one another and divide - a process continuum, perhaps in all organisms. Thus they exhibit polymorphism in their shape and size.
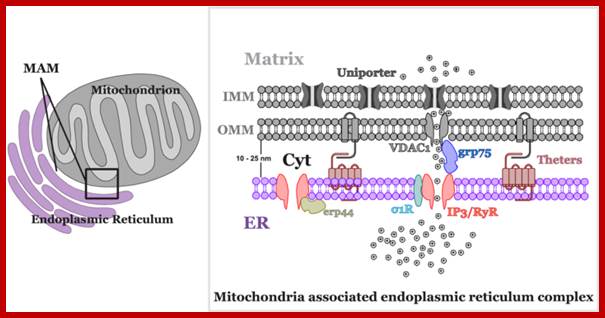
Another, more recent discovery about the structure of mitochondria is the close association that they can have with the endoplasmic reticulum (ER), called the mitochondria-associated ER membrane or MAM. Mitochondria store signaling molecules like Ca2+; Mitochondria can also participate in cell signaling through the reactive oxygen species (ROS) that are created during metabolism. http://frombenchtobedside.wordpress.com/
Communication with each other in terms of chemicals they exchange: www.glogster.com
Ivan Jaijic et al;http://www.mdpi.com/
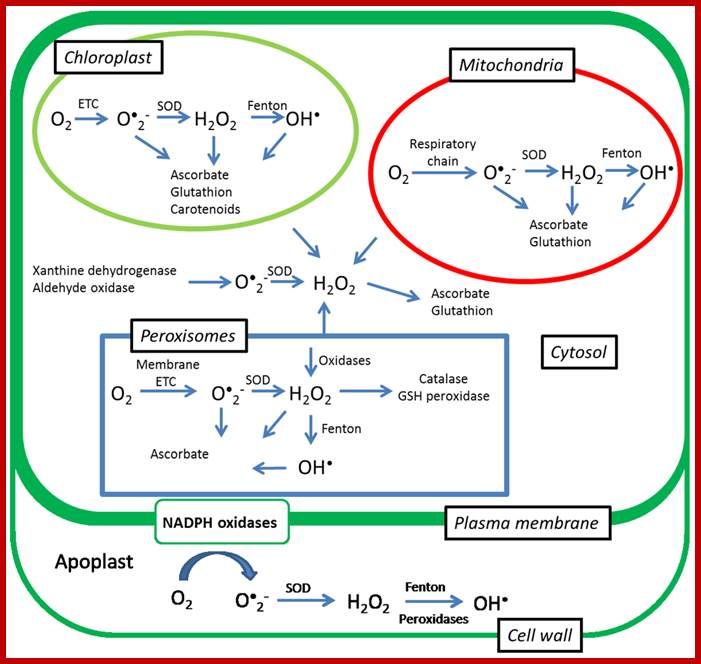
Senescence, Stress, and Reactive Oxygen Species:
Generation of reactive oxygen species (ROS) is one of the earliest responses of plant cells to various biotic and abiotic stresses. ROS are capable of inducing cellular damage by oxidation of proteins, inactivation of enzymes, alterations in the gene expression, and decomposition of biomembranes. On the other hand, they also have a signaling role and changes in production of ROS can act as signals that change the transcription of genes that favor the acclimation of plants to abiotic stresses. Among the ROS, it is believed that H2O2causes the largest changes in the levels of gene expression in plants. A wide range of plant responses has been found to be triggered by H2O2 such as acclimation to drought, photooxidative stress, and induction of senescence. Our knowledge on signaling roles of singlet oxygen (1O2) has been limited by its short lifetime, but recent experiments with a flumutant demonstrated that singlet oxygen does not act primarily as a toxin but rather as a signal that activates several stress-response pathways. In this review we summarize the latest progress on the signaling roles of ROS during senescence and abiotic stresses and we give a short overview of the methods that can be used for their assessment. Ivan Jaijic et al;http://www.mdpi.com/
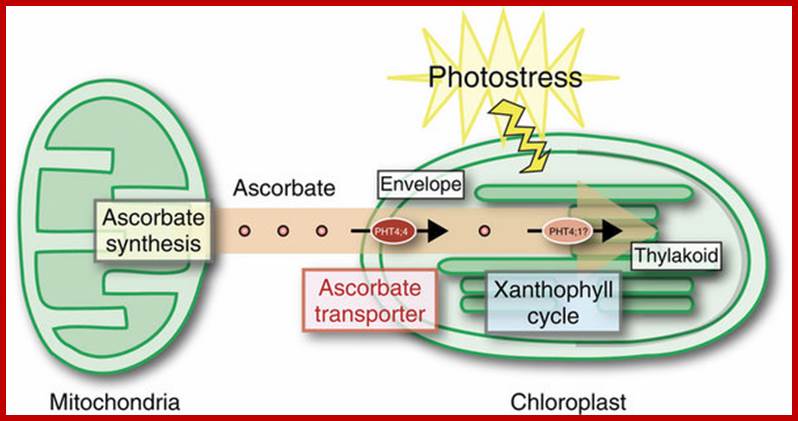
Schematic model of ascorbate transport in chloroplasts.
Upon photostress, PHT4;4 gene expression is enhanced, and the PHT4;4protein at the envelope membranes takes up ascorbate from mitochondria, which is transferred into the thylakoid through an as yet unknown transporter. PHT4;1 is a candidate ascorbate transporter at the thylakoid membrane; Takaaki Miyaji et al. http://www.nature.com/
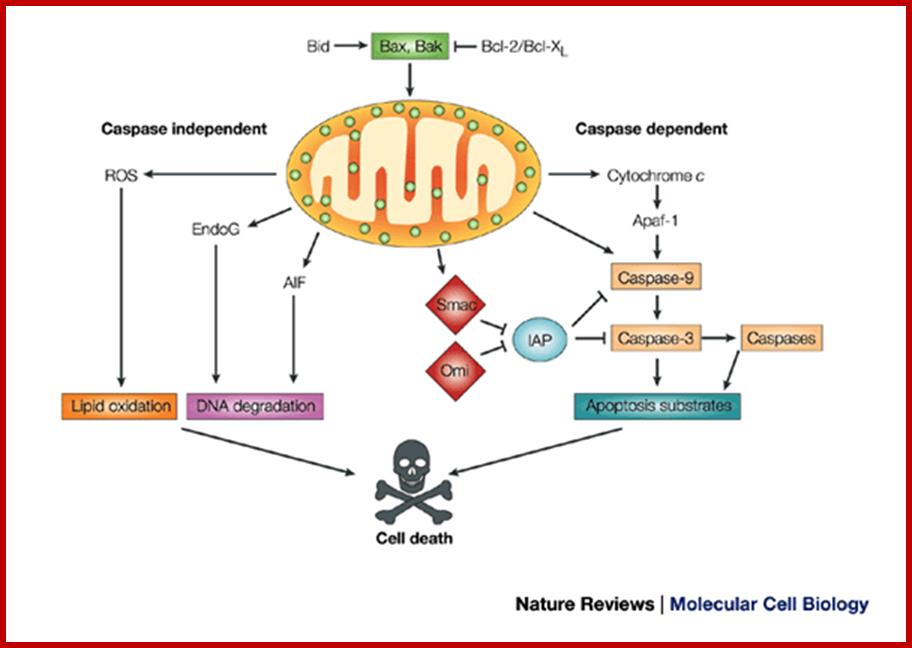
Mitochondria are involved in regulating the cell cycle and play a key role in the apoptosis pathway, a type of programmed cell death. And just for the sake of completeness: some parts of the steroid hormone, heme synthesis and urea cycle pathways take place in mitochondria. http://frombenchtobedside.wordpress.com/
Number and Distribution:
The number of mitochondria found in cells varies from cell type to cell type; it varies from species to species. Even physiological state of the cell determines the number. Trypanosome cells contain only one mitochondrium. Yeast cells under glucose repression posses just one or two mitochondria per cell. But a liver or meristems in contain 500-1600 mitochondria per cell. In some amphibian oocytes which are very active, there can be 10,0000 or more mitochondria. The cell that requires more energy contains more number of mitochondria, ex: flight muscle cells in insects, heart cells have more mitochondria than their intestinal cells.
Even the intracellular distribution and orientation of mitochondria depends upon the structures involved in a particulars function. For example, in flight muscle cells, in the vicinity of centrosomes, mitochondria are arranged radially and mitochondria are packed longitudinally. At the basal granules of flagella they are aggregated at the base; otherwise they are randomly distributed in the cytoplasm. Mitochondrial dysfunction can cause aging, and found to operate in diabetes 2 and Alzheimer’s disease
Structure:
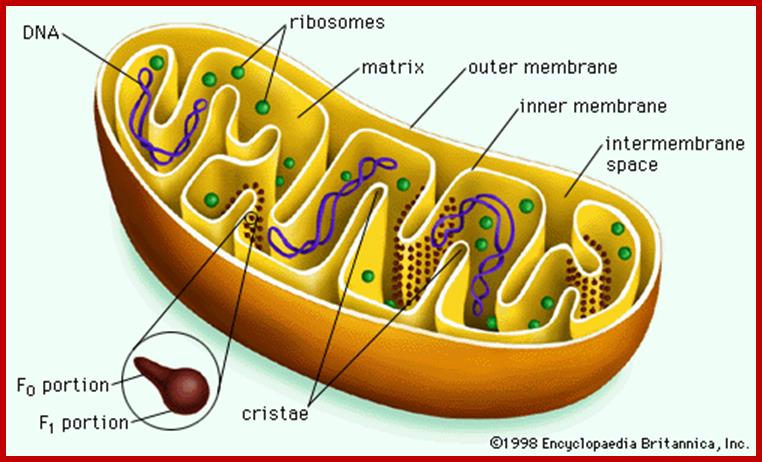
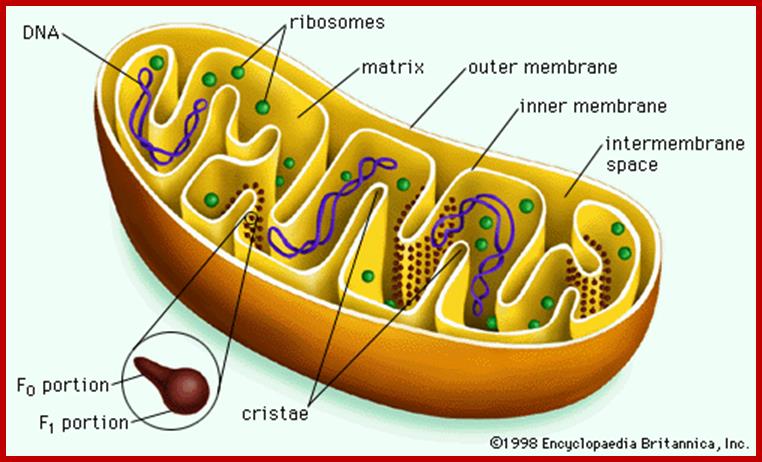
Probably mitochondrion is the only cell organelle whose structure has been studied in greater details. Like any other cell organelles, it is also bounded by membranes. The outer membrane is 6 nm thick and the length varies from mitochondria to mitochondria and from to cell. Structurally mitochondrion consists of two membranes, the outer and the inner membranes and they are separated from each other with perimitochondrial space. The outer membrane can be striped of from the inner membrane by dgitonin treatment and such mitochondria are called mitoplasts.
The outer membrane can be easily identified by its marker enzyme called monoamine oxidase. The outer surface of the outer membrane appears to be smooth but careful observations reveal the presence of many aggregated granular structures which are believed to be enzymes responsible for glycolytic process. The perimitochondrial space is filled with a fluid in which, enzymes like Adenosyl cyclases Cyt.c and others are found.
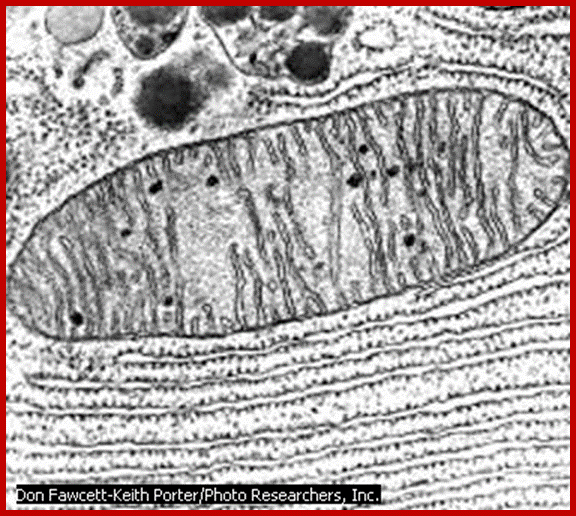
The inner membrane shows structural complexity which is unique to this organelle. It is folded into a number of membranous crests called cristae. They may be oriented transversely or longitudinally. But the number of cristae varies and it depends upon the functional state of the cell. For example, mitochondria found in flight muscle cells of insects contain 10,000 or more cristae, but in resting cells the number of cristae in mitochondria is few. The increase in the number of cristae increases the surface area of reaction which helps in the production of more of ATPs.
Though cristae provide a kind of compartmentation to mitochondrial chamber, it is not total. The perimitochondrial space is continuous in the cristae folds. In spite of both membranes being distinct, they are in contact with each other at least at 70-100 sites. Recent studies, involving mitochondrial contraction and expansion due to changes in energy levels of mitochondria, have demonstrated that most of the mitochondria exhibit several contact zones with ER; where at the cytoplasmic surface a cluster of ribosomal complexes are found. It is speculated that it is through this region that some of polypeptides synthesized in the cytosol are transported into mitochondia. Probably these regions are specialized structures involved in the transportation of components from cytoplasm into mitochondria. Now it is known that there are specific transporters of nuclear coded proteins are transported into mitochondria through such transporters. The outer membrane has Tom and the inner membrane has Tim. These have been characterized. Basically these contain a receptor protein and channel proteins.
The inner surface of the cristae membranes ie, towards mitochondrial matrix side, is studded with a large number of knob or globular shaped particles with a conspicuous stalk like structure. Such globular particulate are called Racker particles which have been identified as F1 particles. They act as ATP synthetases and they are uniformly spaced at 10 nm apart. The number of F1 particles present in each mitochondria ranges from 100 to 1000. The cristae membranes also contain a large number of electron transporting enzymes and other oxido reductases deeply buried in the core of the lipid bilayers, but vectorially organized. In fact, electron transporting enzymatic units constitute more than 20% of the total membrane proteins.
Mitochondrial chamber is filled with fluid called mitochondrial matrix. A host of enzymes are present in this fluid. The unique feature of matrix is the presence of a circular DNA duplex and all t RNAs, and mRNAs. It also contains 70s ribosomes which are similar to that of prokaryotes. Having the above said structures, mitochondrial is considered as a semiautonomous organelle.
Chemical Composition and Functions:
The outer membrane consists of 40% lipids and 60 percent proteins. The common liquids found are lecithin, cephalin and cholesterol. Among proteins, along with structural ones, the outer membrane posses a marker enzyme called monoamine oxidase. On the contrary, the inner membrane is made up of 20% lipids and 80% proteins. The notable lipids present are cholesterol, lecithin and cephalin and cardiolipins, the latter is present is significant amount. Among proteins present in the inner membrane are ATP synthetase succinic dehydrogenase carrier proteins like carnitine, fatty acid acyl transferase, ATP and ADP carrier, which transport inorganic phosphates, calcium, malate, glutamate, and many others.
The electron transport enzymes, proton secreting proteins are virtually buried in the core of the inner membranes. However, electron transporting proteins, which include oxido-reductases, are grouped into four complexes. Each of them vectorially arranged for receiving the reduced coenzymes like FADH2 and NADH2 which are then subjected to terminal oxidation, while the outer is virtually impermeable even for simple ions. They have to be transported across the membrane through specific carriers.
Mitochondrial matrix also consists of a wide variety of enzymes for kreb’s cycle, amino acid metabolism, protein metabolism, nucleic acid metabolism and fatty acid oxidation.
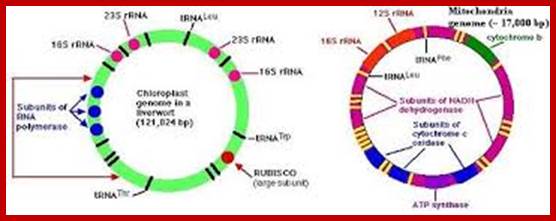
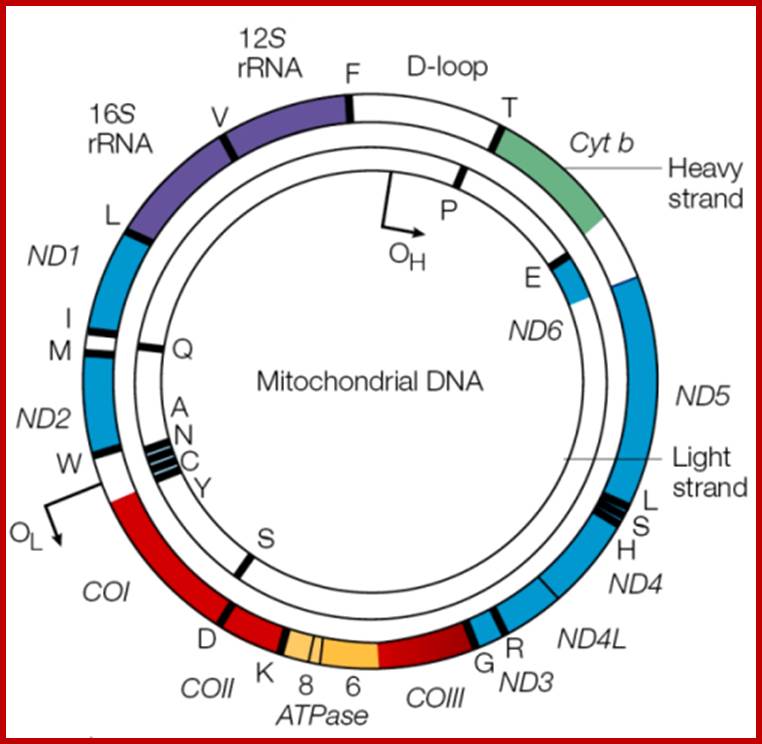
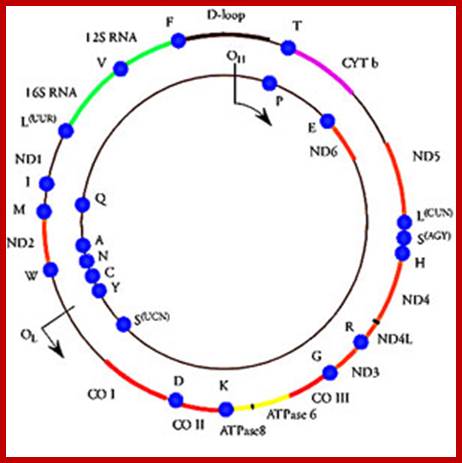
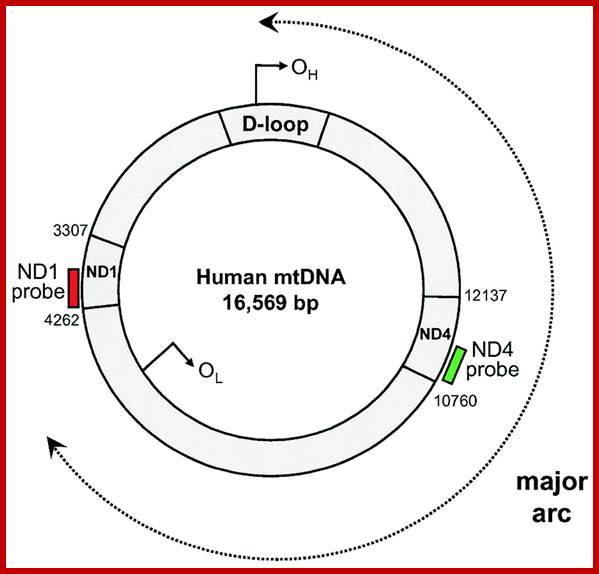
Mitochondrial DNA found in the matrix is found in multiple copies from 40 to 60 or more. The DNA is in circular form and codes for 2rRNAs, all 22tRNAs and 13 mitochondrial proteins. Mammalian mitochondria of human contains a circular DNA of ~16500 bases pairs, but yeast mit is 2-3 times larger than mammalian and it has been mapped. Among 13 proteins coding genes, three for 3 large polypeptides of cytochrome oxidase, and one for the large subunit of cyt.B-c reductase and one hydrophobic protein “transfer protein” of ATPase complex are present. The other gene products which have not been identified are called URF1, URF2 etc. The unique feature of mit DNA is that most of the gene products are transcribed on H strand, but L strand in also used for the production of transcripts for few tRNAs and one or two URFs. In trypanosome only one mitochondrium is present and it contains Kinetoplastida. It has a large single maxi chromosomal DNA and hundreds of smaller min circular DNAs.
The DNA replication in mitochondria is independent or sometime synchronized with that of nuclear DNA replication. But the mechanism of replication, though it is semiconservative it exhibits D-loop mechanism. Another important feature of mit DNA is all genes are tightly packed with small spacers or intervening sequences in between the genes or within the transcribing genes. tRNA genes are located in between protein coding frames. However, yeast mitochondria have a long DNA with redundant and intervening sequences within the coding sequences. In plants, mitochondrial DNAs, besides having the above said genes they also have a gene for male sterility, ex: Zea mays.
Mitochondrial Translational Machinery:
The ribosomal machinery present in mitochondrial matrix is typical of prokaryotic 70s type, but in some plants the ribosomes presents in mitochondria are slightly smaller than 70s. Nevertheless the 23s RNA, 16s rRNA and 5s RNA are present with some minor differences. Most of the transcribing the translational factors and other enzymes found in mitochondrial matrix are coded for by nuclear genome and translated on cytosol 80s ribosomes, then they are imported into mitochondria. From the ribosomes it is clear that mitochondrial matrix has all the components for nucleic acid & protein synthesis.
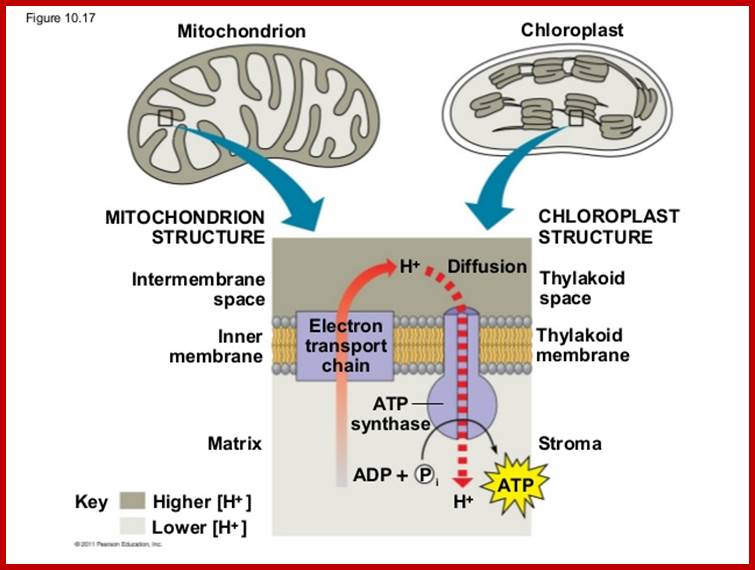
Mitochondrial matrix is also involved in citric acid cycle, where energy rich co-enzymes like FADH2 and NADH2 are produced. They are then taken up by cristae membranes, where they are step wisely and sequentially oxidized. During this process, the energy released is used for the synthesis of ATP. This phenomenon is called oxidative phosphorylation. These mitochondria are endowed with a unique structural and functional co-ordination.
Mitochondrial Genome:

Organization of the mammalian mitochondrial genome. Thirteen protein-coding genes (yellow), twenty-two tRNA genes (red) and two rRNA genes (orange) are encoded on a single circular nucleic acid and transcribed from three promoters (blue): LSP, HSP1 and HSP2, which are situated in a single region called the D-loop, which contains regulatory sequences that control transcription from all three promoters, including motifs for DNA-binding proteins such as Tfam. The inner circle of genes is encoded on the (-) strand and transcribed from the LSP promoter. The outer circle of genes is encoded on the (+) strand and transcribed from the HSP1 and HSP2 promoters. Transcription from HSP2 is terminated distal to the 16S rRNA gene. The resulting three polycistronic transcripts are processed by enzymatic excision of the tRNAs (red). ATP6, ATP8, subunits of ATP synthase F0; Cox1, Cox2, Cox3, subunits of cytochrome oxidase; CytB, cytochrome B, Nd1, Nd2, Nd3, Nd4, Nd4L, Nd5, Nd6, subunits of NADH dehydrogenase. http://www.genomebiology.com/
Heidi Chial, Ph.D. ;www.nature.com
· Human Mit DNA is circular and 16,569bp long; Mit DNA contains 37 genes, that codes for 13 proteins, 22 tRNAs and 2 rRNAs; The mitochondrial genome is circular, whereas the nuclear genome is linear (Figure above).
· The mitochondrial genome is built of 16,569 DNA base pairs, whereas the nuclear genome is made of ~3.3 billion DNA base pairs.
· The mitochondrial genome contains 37 genes that encode 13 proteins, 22 tRNAs, and 2 rRNAs.
· The 13 mitochondrial gene-encoded proteins all instruct cells to produce protein subunits of the enzyme complexes of the oxidative phosphorylation system, which enables mitochondria to act as the powerhouses of our cells.
· The small mitochondrial genome is not able to independently produce all of the proteins needed for functionality; thus, mitochondria rely heavily on imported nuclear gene products.
· One mitochondrion contains dozens of copies of its mitochondrial genome. In addition, each cell contains numerous mitochondria. Therefore, a given cell can contain several thousand copies of its mitochondrial genome, but only one copy of its nuclear genome.
· The mitochondrial genome is not enveloped, and is it not packaged into chromatin.
· The mitochondrial genome contains few, if any, noncoding DNA sequences. (Three percent of the mitochondrial genome is noncoding DNA, whereas 93% of the nuclear genome is noncoding DNA).
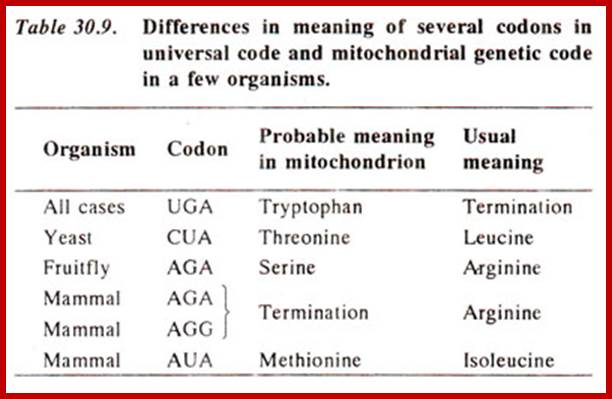
· Some mitochondrial coding sequences (triplet codons) do not follow the universal codon usage rules when they are translated into proteins.
· Some mitochondrial nucleotide bases exhibit functional overlap between two genes; in other words, the same nucleotide can sometimes function as both the last base of one gene and the first base of the next gene.
· The mitochondrial mode of inheritance is strictly maternal, whereas nuclear genomes are inherited equally from both parents. Therefore, mitochondria-associated disease mutations are also always inherited maternally.
· Mitochondrial DNA is coded by both strands, for they transcribe and generate rRNA, tRNA and mRNAs.
Mitochondrial genes on both DNA strands are transcribed in a polycistronic manner: Large mitochondrial mRNAs contain the instructions to build many different proteins, which are encoded one after the next along the mRNA. In contrast, nuclear genes are usually transcribed one at a time from their own mRNA. http://www.nature.com/
Mitochondrial mutations can lead to aging and cancer. Mitochondrial disease are many such as Leber hereditary optic neuropathy (LHON), causes postlingual deafness (deafness that occurs after three years of age, when a child has already learned to speak), diabetes, examples include Pearson syndrome, Leigh syndrome, progressive external ophthalmoplegia, exercise-induced muscle pain, fatigue, and rhabdomyolysis and many others.
Biogenesis:
In the past 20-30 years various speculative theories have been proposed from time to time to explain mitochondrial biogenesis. Most of the views were based on cytological observations. But now the structure, genetic composition and its functional aspects are fairly known. Recent studies clearly indicate that mitochondria originate from the pre existing mitochondria by growth and fission. The biogenesis of mitochondria is highly regulated and co-ordinates with the nuclear genome. Though, more than 95% of the total mitochondrial proteins are coded for by nuclear genome but certain factors coded for by the nuclear genome exert control over mitochondria gene expression.
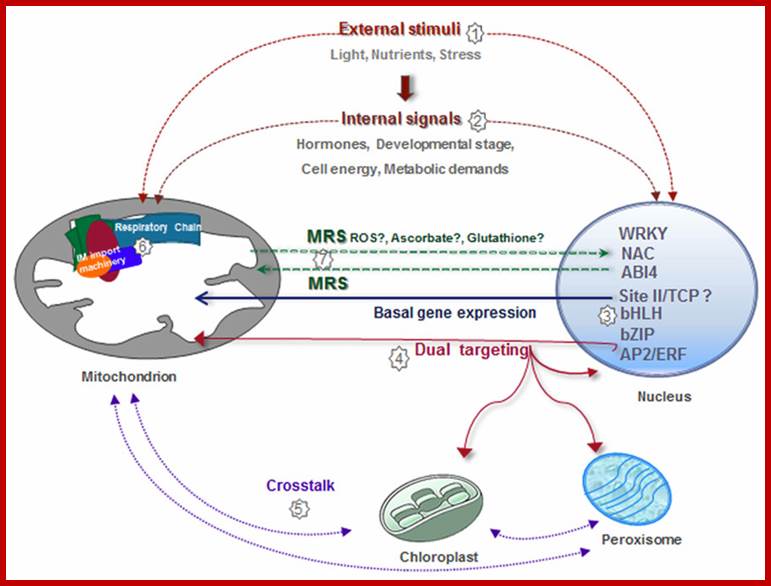
Coordination of plant mitochondrial biogenesis: keeping pace with cellular requirements ;Elina Welchen etal, http://journal.frontiersin.org/
Similarly, some mitochondrial protein factors coded for by the mitochondrial genes are transported into nucleus where they regulate the expression of genes and gene products required for mitochondria (one report from Neurospora). In this coordinated network, mitochondria need to generate signals to modify nuclear gene expression according to organelle and cellular requirements. Signaling pathways from organelles to the nucleus are referred to as Retrograde Signaling pathways. While several relevant players in chloroplast retrograde signaling have been extensively characterized (for reviews see Leister et al., 2011;Kleine and Leister, 2013), relatively little is known about mitochondrial retrograde signaling (MRS). Considering that chloroplasts and mitochondria are closely connected, signaling pathways involved in chloroplast retrograde regulation need to be explored in the context of mitochondria to get new insights into MRS.
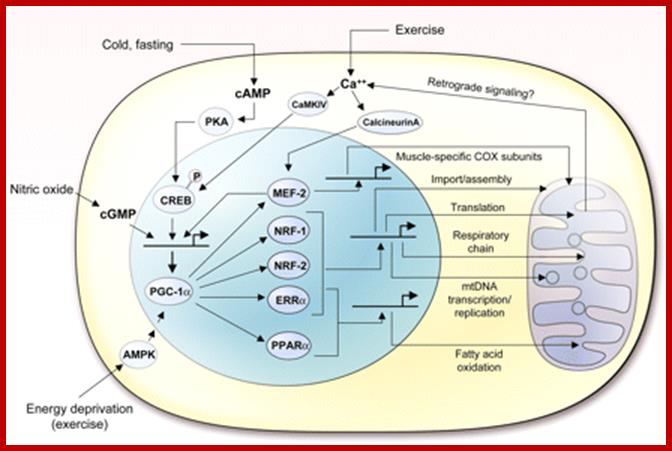
PGC1-alpha mediated pathway governing mitochondrial biogenesis and function; certain transcription factors NRF-1, NRF-2, ERRa, PPARa and MEF-2 act on PGC-1aloha; http://physrev.physiology.org/
As mitochondrial genome and 70s ribosomal machinery and other transcription translating factors are more or less similar to that of prokaryotic bacteria, it is viewed by many biologists that the eukaryotic mitochondria were once bacterial cells; in course of time they have incorporated into eukaryotic cells properly for ~3 billion years ago. Since then they exist as symbionts. The similarities between bacteria and mitochondria are circular DNA, chloramphenicol sensitive,70s ribosomes, 23s RNA, 16s RNA and 5s RNA. Based on these observations, it is now strongly believed that mitochondria are symbiotic bacteria; now it is an accepted fact. Mitochondrial codons are not universal. Mitochondrial DNA has been used to find out phylogeny. It is now believed all the present 6 billion humans have derived mitochondria from few African mothers. Mitochondrial DNA gives foot print of human race; for example 4.5000 to 5000 years ago human population lived around Indus valley, are now called with the arrival of Aryans (now called Brahmins, most of them from Iran called Hindus). Today take all casts and creeds of human population in India, if one observes mit. DNA one finds this Indus valley lived women for only women contribute mitochondria to their offspring’s. The origin of humans took place in Africa 120,000-165000 years ago, this population moved to India via Indus valley around 60,000 years ago, and this population moved to China and Australia 50k and Europe around 40K ago. Indian women have its imprint in the form of Mitochondrial DNA, which provided inheritance pattern that we see in human population whatever cast or communities you belong. You cannot question the poof of DNA. You cannot question this genetic inheritance, so also Male Y chromosomal DNA.
Prof. Aleksandra Trifunovic; www.cecad.uni-koeln.de
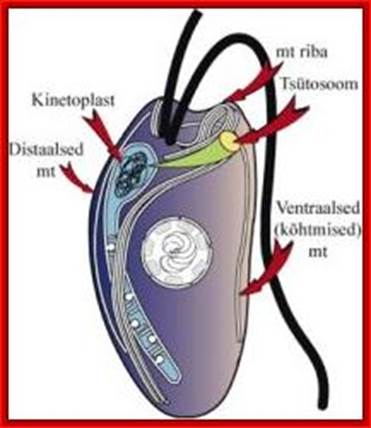
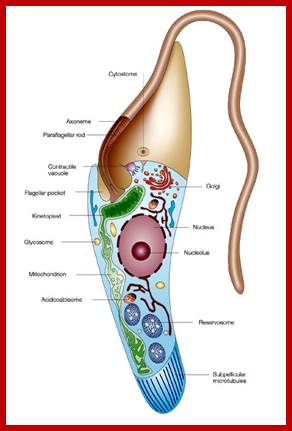
Kinetoplastida; https://en.wikipedia.org; http://www.nature.com/
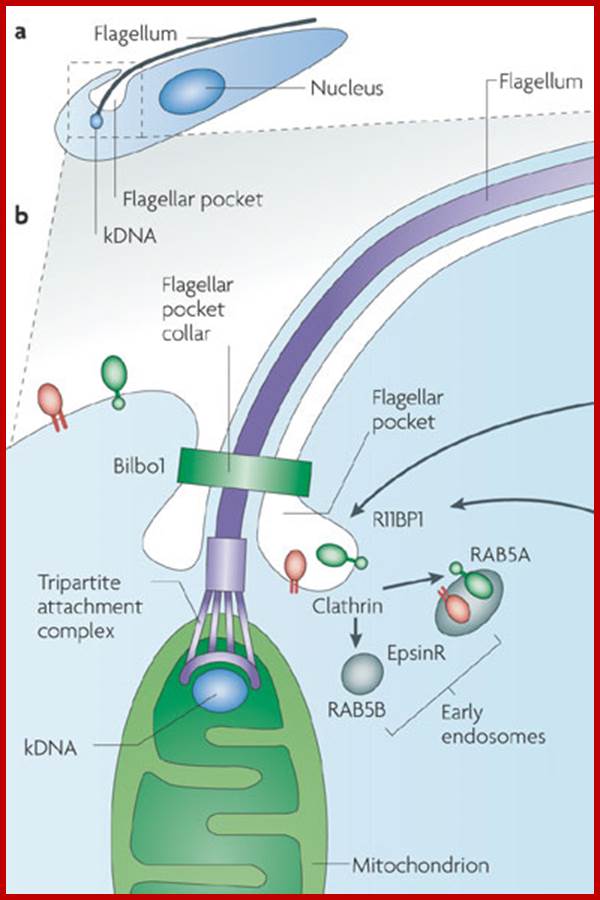
Kinetoplastid DNA (kDNA) is organized into an incredible network of interlocked rings. The kDNA codes for cytochrome b oxidase subunits, NADH dehydrogenase subunits, ATPase subunit 6 and rRNAs By studying this amazing structure, how did scientists learn about RNA editing?
Laura Vargas-Parada,2010 nature.; www.nature.com
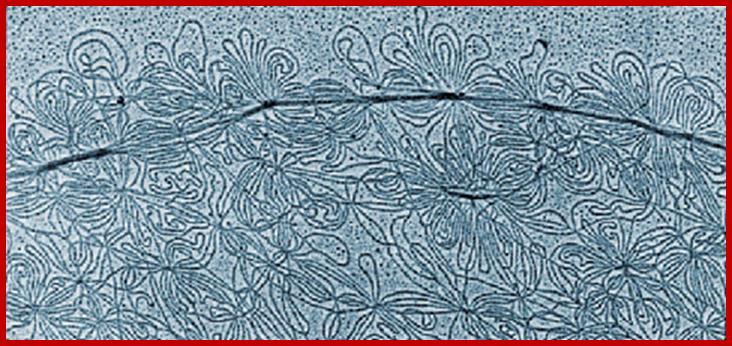
Concatenated DNA of the Kinetoplast of kinetoplastids (Trypanosoma species and Leishmania species etc).
Very interesting is the kinetoplastid, this structure is present in Trypanosoma cruzi, T. rhodesience, T.gambiense, T.brucei and other similar trypanosomes.. It contains a single mitochondrium and dense granule within its mitochondria. This structure consists of concatenated circular DNA and they are associated with structural proteins and RNA polymerases. This kinetoplastid is found at the base of the flagella. The DNA is very unusual for it has two different DNA, one maxicircle and another minicircular DNAs catenated to each other. Each network consists of 40-50 maxicircles and 5000-10,000 minicircles. Te maxicircle DNA can be as long as 20-40kb. The minicircle DNA is about 645bp to 2500bp long. In some like L. tarantole maxicircle DNA include guide RNA (gRNA) genes whose transcripts (thousands) associated with proteins (editosomes) perform editing mRNAs either deleting or addition of Uridine.
Maxicircle DNA encodes for proteins needed for mitochondrial function. Minicircle DNA encode for guide RNAs which encode decode the encrypted maxicircle information. Reproduction of this kinetoplastid DNA takes place by disconnecting these rings from the parental kinetoplast DNA and subsequently join with the daughter Kinetoplast DNA.
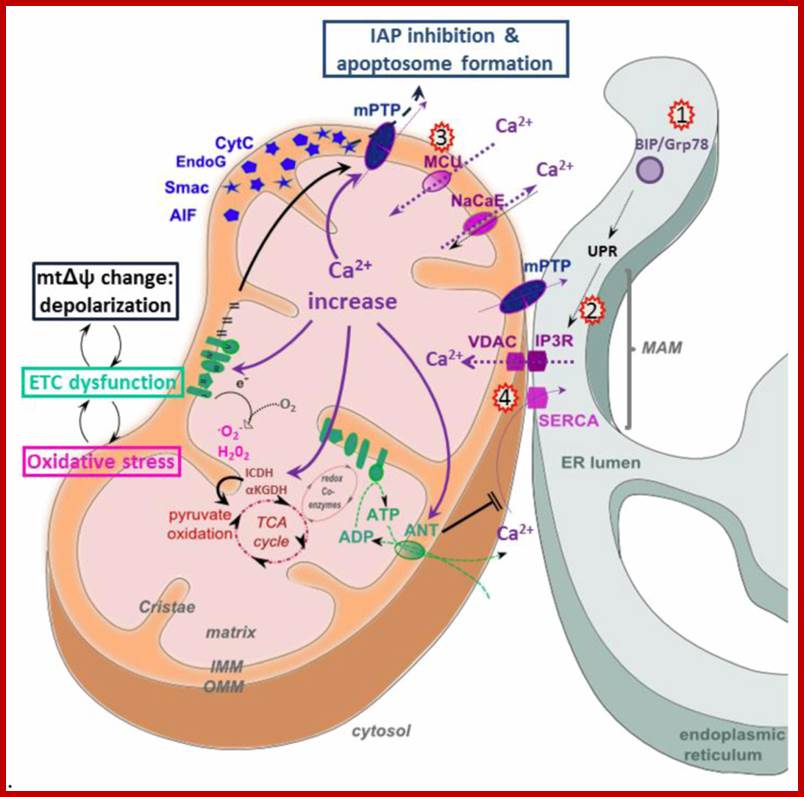
Mitochondria very often found associated with smooth ER andCa2+ is transported into mitochondria. www.mdpi.com;

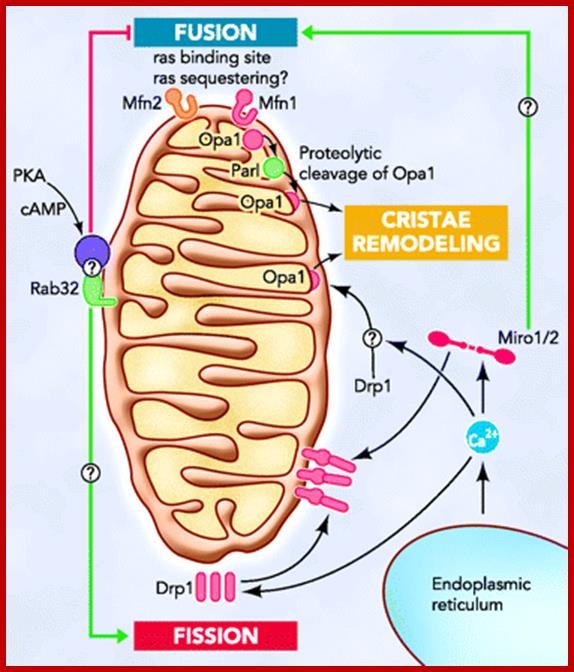
Hypothetical models of the role of contacts between mitochondria and ER in apoptosis; The hFis1/Bap31 platform transmits the mitochondrial stress signal to the ER via the activation of procaspase-8. The cytosolic region of the ER integral membrane protein Bap31 is cleaved by activated caspase-8 to generate proapoptotic p20Bap31, which causes rapid transmission of ER calcium signals to the mitochondria via the IP3 receptor. At close ER-mitochondria contact sites, mitochondria takes up calcium into the matrix via the mitochondrial calcium channels MICU1 or LETM1. The massive influx of calcium leads to mitochondrial fission, cristae remodeling, and cytochrome release. Mfn2 is enriched in the mitochondria-associated membranes (MAM) of the endoplasmic reticulum (ER), where it interacts with Mfn1 and Mfn2 on the mitochondria to form inter-organellar bridges. Upon apoptosis signal, a BH3-only member of the Bcl-2 family, Bik, induces Ca2+ release from the ER and, in turn, induces Drp1 recruitment to the mitochondria and their fragmentation and cristae remodeling. SERCA- sarco /endoplasmic reticulum Ca2+-ATPase. MICU1, mitochondrial calcium uptake1. LETM1,leucine zipper/EF hand-containing transmembrane 1. http://www.hindawi.com/
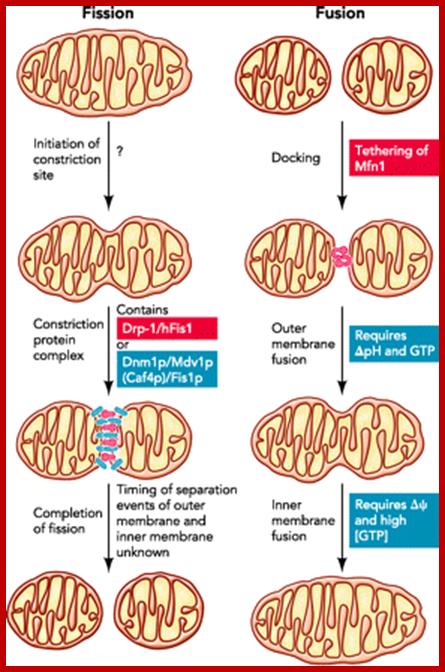
Steps of mitochondrial fission and fusion; The figure highlights our current knowledge of the mechanisms of fission and fusion in yeast (blue) and mammalian cells (red). For discussion, see text;h ttp://physiologyonline.physiology.org/
Fission: The current model of fission in yeast is based on a trimer complex of Dnm1p, Mdv1p, and Fis1p . The current model supports that Dnm1p translocates on activation to mitochondria, where it functions as a mechanoenzyme constricting mitochondrial membranes like its homolog dynamin I constricts the nascent endocytotic vescicle. In one model, a stable Fis1p-Mdv1p complex is essential to recruit Dnm1p, which then homo-oligomerizes. In a second model, Dnm1p localization to mitochondria is independent of Fis1p and Mdv1p, which are conversely responsible for the Dnm1p oligomerization step (FIGURE 2⇓). The protein Caf4p seems to have a similar adaptor function as Mdv1p. How mitochondrial fission is regulated remains obscure. Remarkably, blockage of fission is not lethal to yeast, suggesting that mitochondria can still divide during cytokinesis as well as during meiosis and sporulation of diploid yeast cells. Whether this is due to different protein machineries or to mechanical forces remains to be elucidated.
Fusion: Fusion of mitochondria can be divided in at least three steps: docking, fusion of the outer membrane, and fusion of the inner membrane. The trimeric complex of Fzo1p, Ugo1p, and Mgm1p stands in the center of mitochondrial fusion. During docking, two or more Fzo1p on juxtaposed mitochondria interact via their coiled-coil domains. A recently developed in vitro assay showed that fusion of the outer membrane can be separated from that of the inner membrane. Outer membrane fusion requires a pH gradient across the inner membrane and GTP, whereas inner membrane fusion depends on membrane potential and high levels of GTP (79) (FIGURE 2⇑). Whether Mgm1p is responsible for the higher GTP consumption during inner membrane fusion remains unclear. http://physiologyonline.physiology.org/

www.academic.brooklyn.cuny.edu
www.academic.brooklyn.cuny.edu
www.britannica.com; study.com
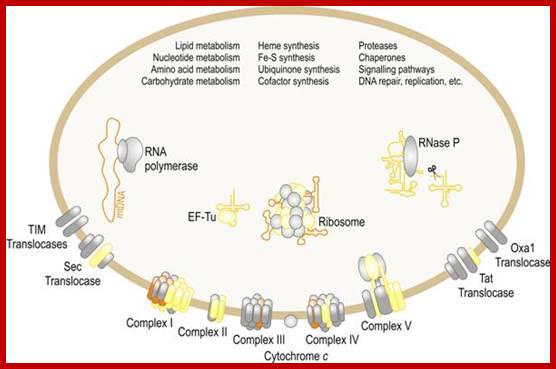
Mitochondria is involved in various functions which are important to cell; www.edoc.hu-berlin.de
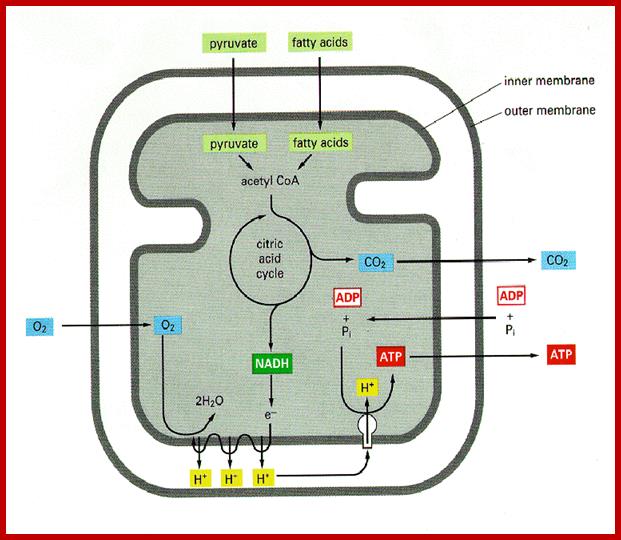
Central role of Kreb’s cycle that takes place in the mitochondrial matrix; www.ridge.icu.ac.jp; www.gopixpic.com
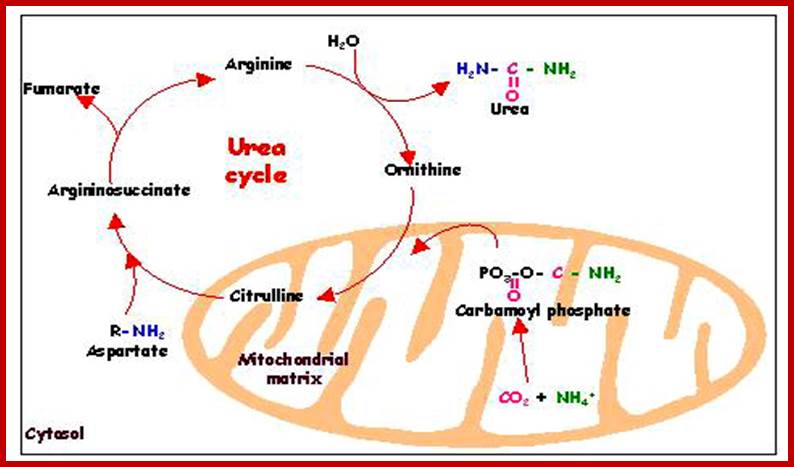
Mitochondria are also involved in urea cycle and below citric acid cycle; edoc.hu-berlin.de
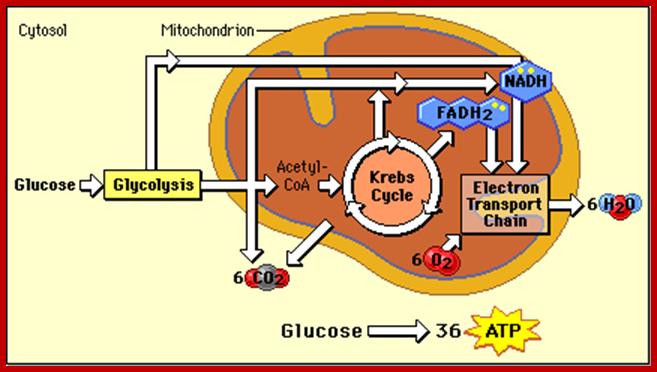
www.nutritionaloncology.org; www.phschool.com
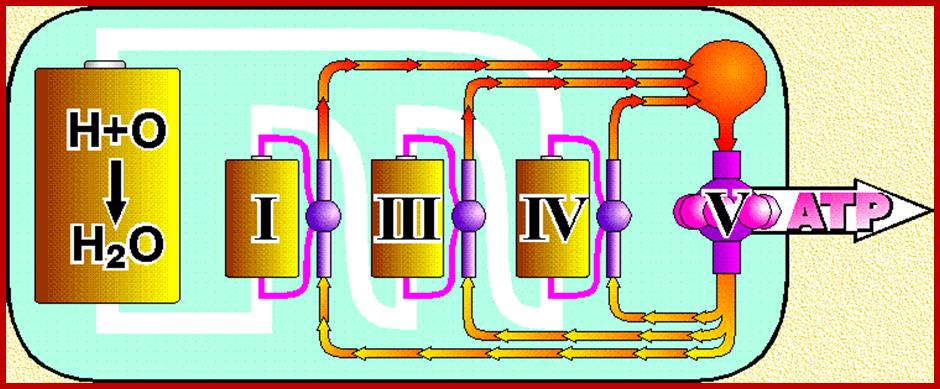
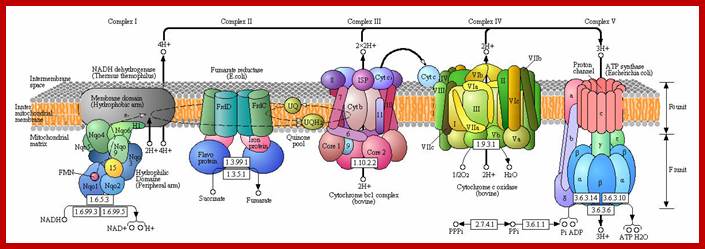
Oxidative phosphorylation- Electron Transport chain
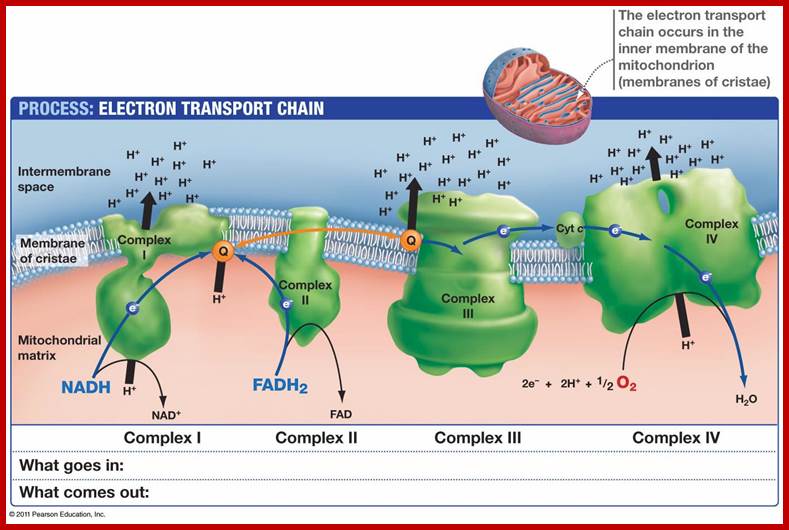
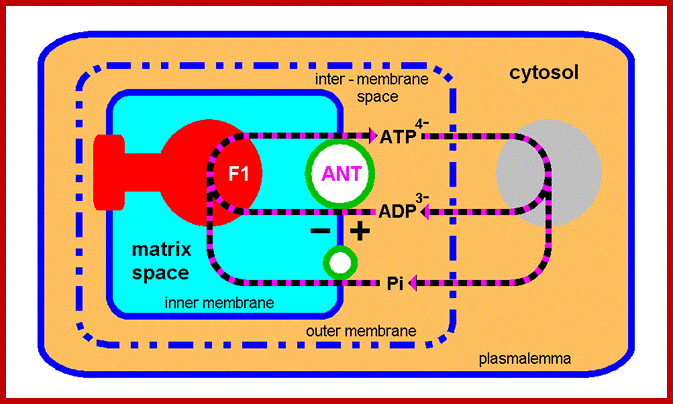
Detailed view of oxidative Phosphorylation; http://employees.csbsju.edu/ biowiki.ucdavis.edu

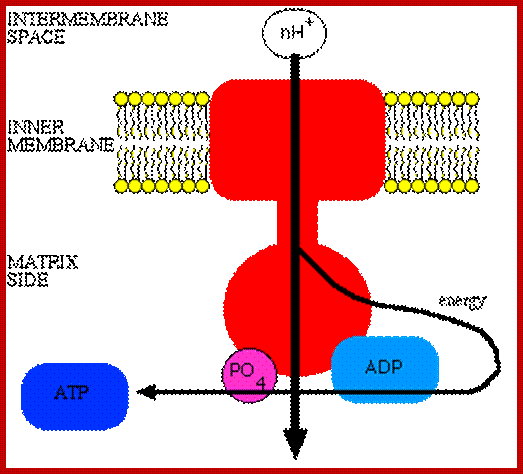
Electron transport chain leads to production of ATP; http://www.exatest.com/ www.ruf.rice.edu

Mitochondrial Genome:
The size of mitochondrial genome varies from one species to the other, human mitochondria contain ~16 Kbp and codes for13 proteins, 22tRNAs and 7 URFs. The number of mit-DNAs per mitochondria can be 100 to 200 and the number of mitochondria per cell varies from one to ten thousand or more. Refer to DNA replication
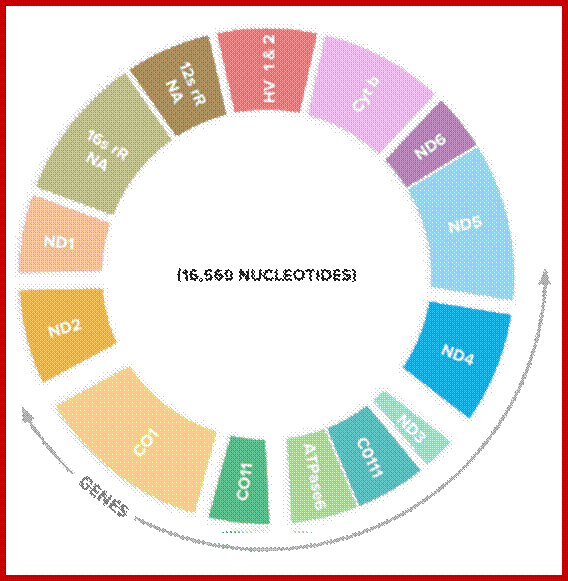 www.genographic.nationalgeographic.com
www.genographic.nationalgeographic.com
Mitochondrial DNA rRNA genes and 13 protein coding genes and ~22 tRNA genes and 6-7 unknown URFs; www.cellbiology.med.unsw.edu.au
Mitochondrial DNA Replication sketch- D-lop mechanism;; www.eplantscience.com; www.nar.oxfordjournals.org

Replication of organelle DNA (Mitochondrial) with Replication Ori-H andOri-L and Trnascriptional start sites-Hsp and Lsp-refer to DNA repication
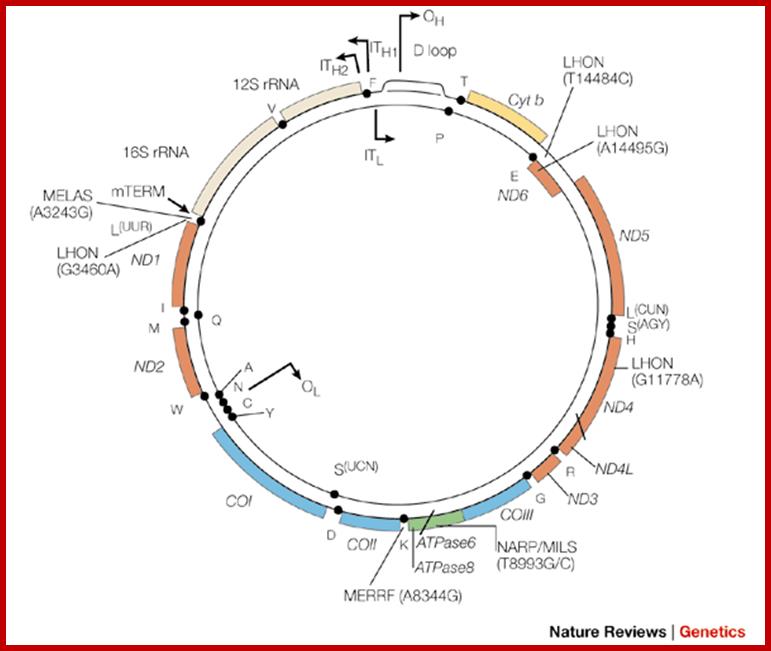
The mitochondrial genome. Jan Smeitink, Lambert van den Heuvel & Salvatore DiMauro; the outer and inner circles represent Havy H and Lighter L strands. The D-loop contain Ori H and Ol on the inner strand. Transcriptional initiation sites are shown as IT- H1 and IT-H2 and IT-L and the direction of transcription isindicated. mTERis mit transcription termination factor binding sites. The 22 mRNA indicated by dots; www.nature.com
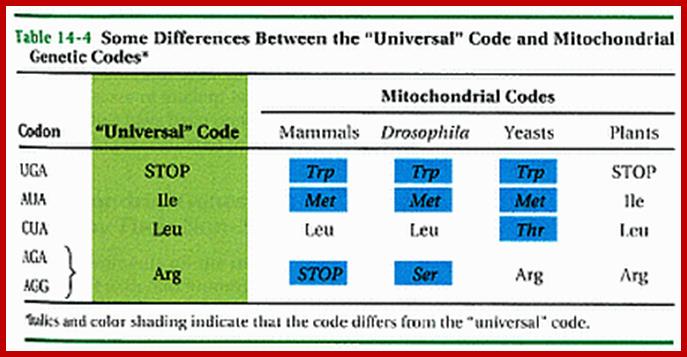
Some genetic codes differ from universal coded, this is specific to mitochondrial codons
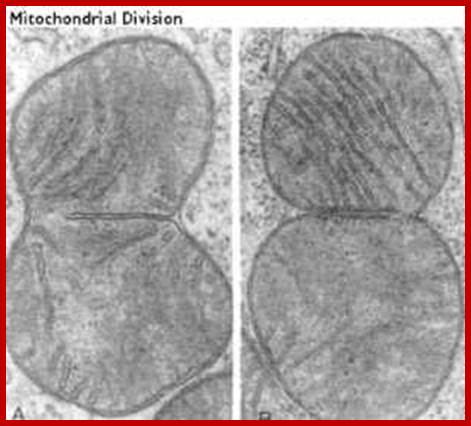
Mitochondrial in the process of division
Mitochondrial Protein Import:
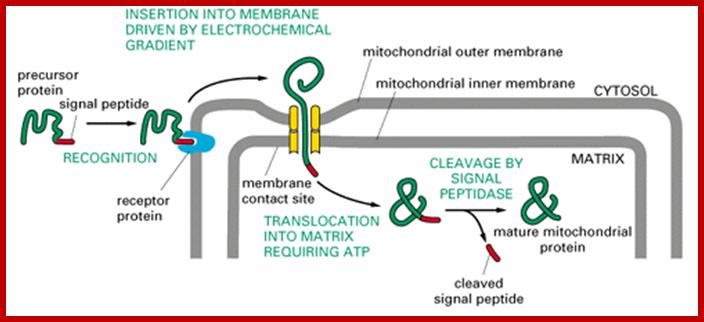
Mitochondria require at least 1000 (yeast)-1500 (human data not consistent among different published articles); proteins for its function, but its genome codes for just 13 proteins in humans an d eight in yeasts and the rest of have to be imported from cytosol. There is intricate communication between mitochondria and nuclear genes with respect to what proteins required by mitochondria and the same are transcribed by the nuclear genome in response to mitochondrial signals; once they are synthesized they are imported. The protein import can function via signals between nucleus and mitochondrial genomes. The import process is two-step process; one transport across the outer membrane and then inner membrane. Interestingly mitochondria are connected to Endoplasmic Reticulum (ER) membranes and it is involved in cellular ion homeostasis and plays an important role in Apoptosis. Though mitochondria inherited from bacteria, majority of the bacterial genes were transferred to host cell nucleus (when?) and they function each contributing to each other. Some of the transporters involed are, TOM proteins (eight), Tim proteins (twelve), SAM three), Pam (two) several others like Mdm, Mda, Hsp, Zim , heat shock proteins and some transporter are export from mitochondria eg; Mss and Pnt1 (export from mitochondria) and few others. Proteins that are imported have N-terminal sequences for the movement of proteins from cytosol into mitochondrial peripheral space that is found between outer and inner mitochondrial membranes. Some of the proteins imported into mitochondrial space are further processed and get inserted to the inner surface of the inner membrane. Many proteins are sorted in inner cristae membranes and some are found in free mitochondrial space.
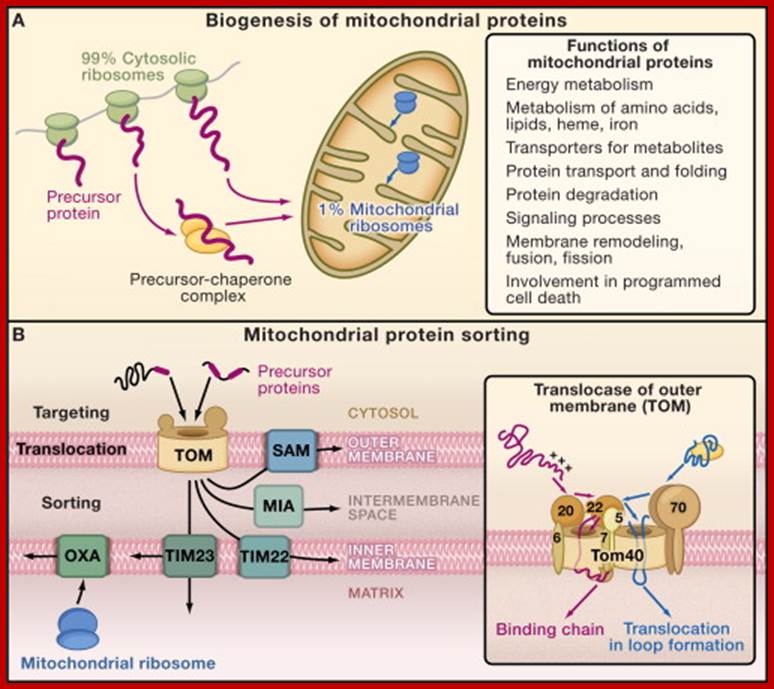
Principles of Mitochondrial Protein Biogenesis; http://www.sciencedirect.com/
(A) Cytosolic and mitochondrial protein synthesis. Most mitochondrial proteins are synthesized on cytosolic ribosomes and are imported into the organelle. About 1% of mitochondrial proteins are synthesized inside the organelle.
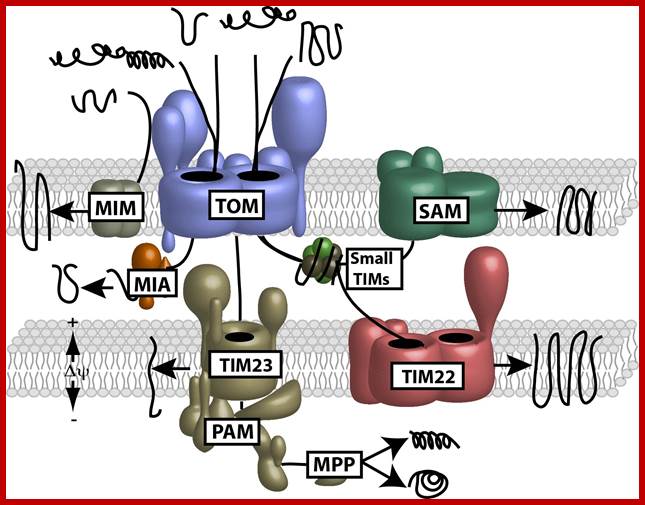
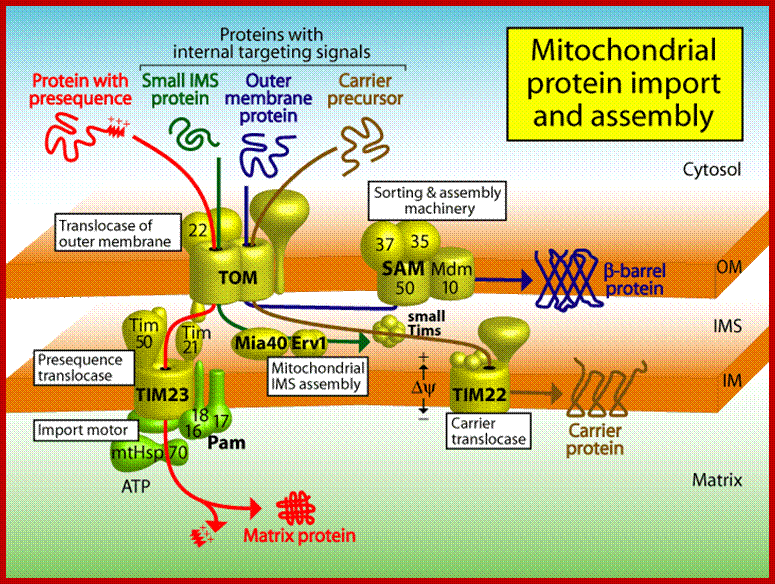
(B) Sorting pathways of mitochondria. The translocase of the outer membrane (TOM complex) is the main entry gate into mitochondria. Subsequently, the precursor proteins follow different sorting pathways. MIA, mitochondrial intermembrane space assembly; OXA, insertase/export machinery of the inner membrane; SAM, sorting and assembly machinery; TIM22 complex, carrier translocase of the inner membrane; TIM23 complex, presequence translocase of the inner membrane. (Inset) The TOM complex consists of seven different subunits. The receptors Tom20, Tom22, and Tom70 recognize precursor proteins and transfer them to the central component, the channel-forming Tom40. Three small Tom proteins, Tom5, Tom6, and Tom7, are involved in the assembly and dynamics of the TOM complex. (Presequence-carrying precursor proteins, red; hydrophobic precursors with internal targeting signals, blue.); http://www.sciencedirect.com/
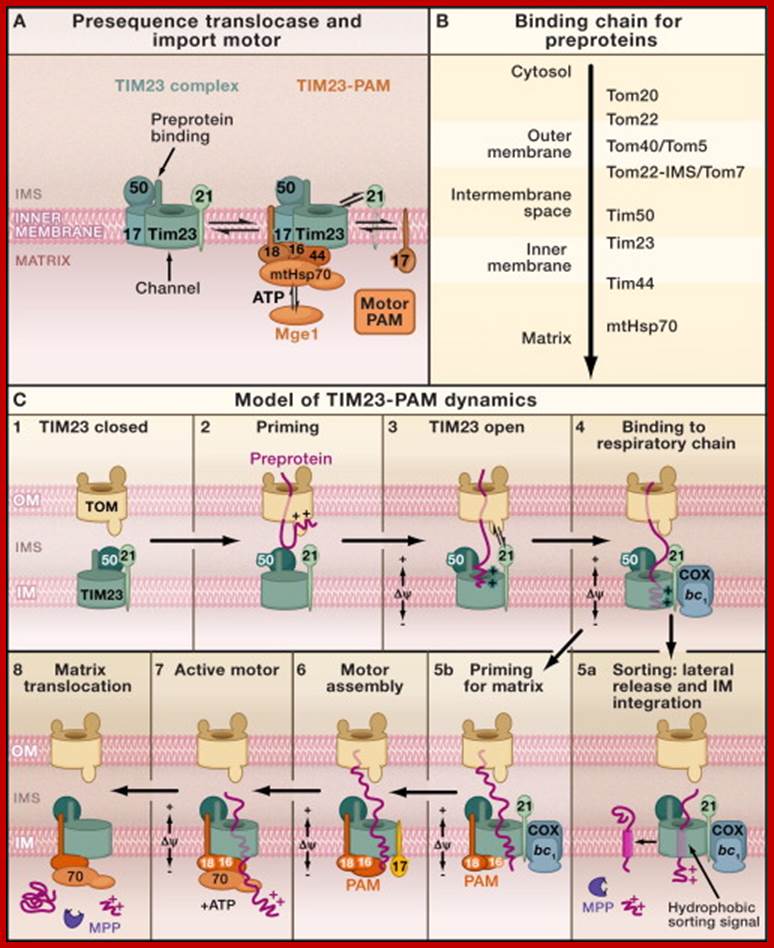
The Presequence Pathway to the Mitochondrial Inner Membrane and Matrix
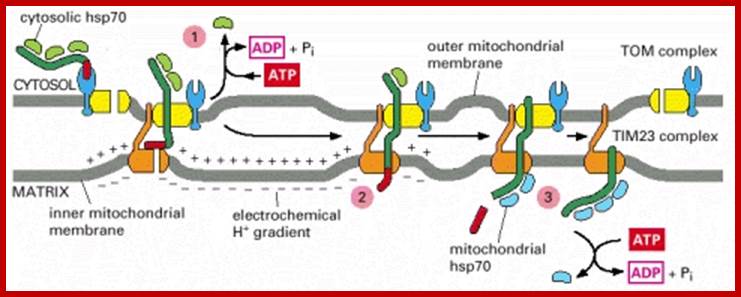
(A) Forms of the presequence translocase of the inner membrane (TIM23 complex). (Left image) Motor-free TIM23 complex that can insert preproteins into the inner membrane. (Right image) TIM23 complex associated with PAM (presequence translocase-associated motor). The central chaperone that binds to preproteins is mtHsp70. The function of mtHsp70 is regulated by the cochaperones Pam16-Pam18, Tim44, Pam17, and the nucleotide exchange factor Mge1. The function of the individual components is described in Table 1.
(B) Presequence-carrying preproteins are directed into the matrix by a sequential chain of binding sites.
(C) Hypothetical model for the dynamic cooperation of the TIM23 complex of the inner membrane (IM) with the TOM complex of the outer membrane (OM), the respiratory chain complexes III (bc1-complex) and IV (cytochrome c oxidase, COX), and the motor PAM. The membrane potential (Δψ) activates the Tim23 channel and exerts an electrophoretic effect on the positively charged presequences of preproteins. ATP drives the action of mtHsp70. The mitochondrial processing peptidase (MPP) removes the presequences.; http://www.sciencedirect.com/
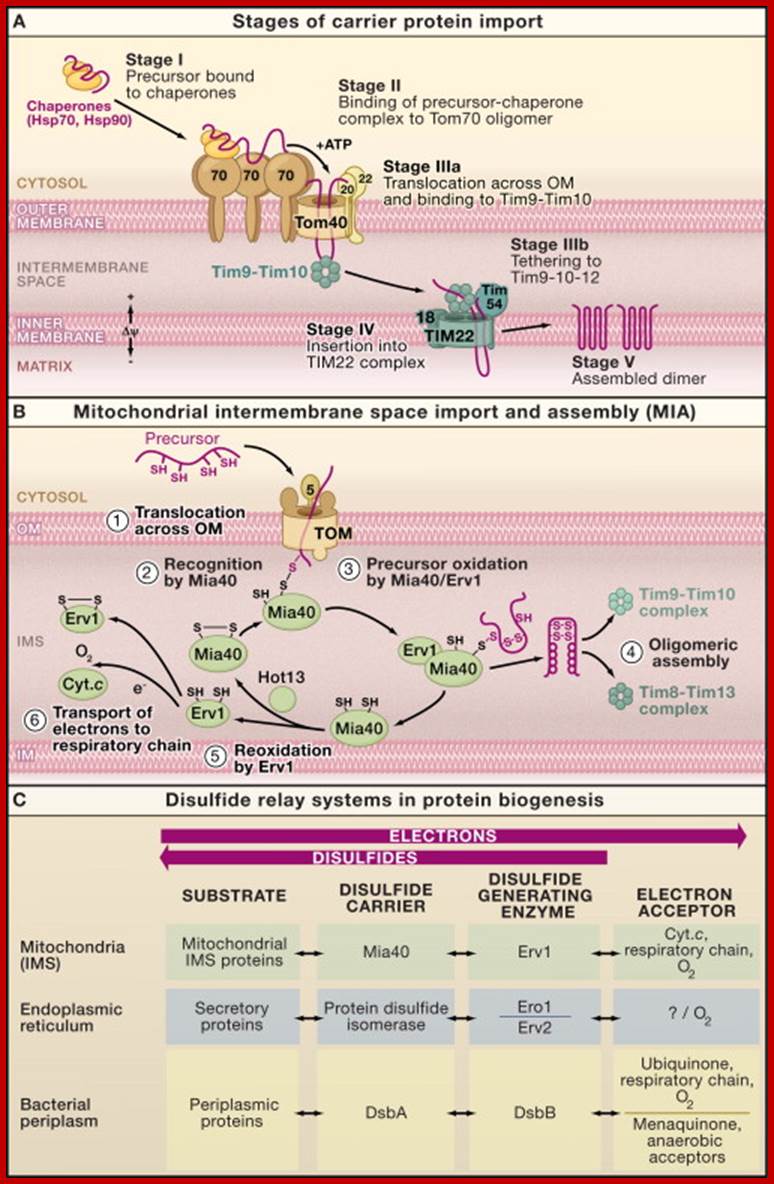
Intermembrane Space Chaperones: The Carrier Pathway, and Machinery for Import and Assembly;
(A) Carrier pathway to the inner mitochondrial membrane. The noncleavable precursors of hydrophobic metabolite carriers of the inner membrane are imported in several stages. Cytosolic chaperones guide the precursor (stage I) to the receptor Tom70 (stage II). The precursor is transported through the translocase of the outer membrane (TOM complex) in a loop formation and interacts with the Tim9-Tim10 chaperone complex (stage IIIa). Tim9-Tim10 guides the precursor through the intermembrane space (IMS) to the carrier translocase of the inner membrane (TIM22 complex with bound Tim9-10-12) (stage IIIb). The membrane potential (Δψ) promotes insertion of the precursor into the inner membrane via the TIM22 complex (stage IV), followed by assembly into the mature form of the carrier protein (stage V).
(B) The machinery for import and assembly (MIA) of preproteins in the mitochondrial intermembrane space is required for small IMS proteins with cysteine motifs. The IMS receptor Mia40 binds to the precursor via a transient disulfide bond. The sulfhydryl oxidase Erv1 cooperates with Mia40 in the oxidation of the precursor protein. Erv1 reoxidizes Mia40 and transfers electrons to cytochrome c (Cyt. c). A third component, Hot13, assists in the oxidation of Mia40.
(C) Comparison of the disulfide relay systems of the mitochondrial intermembrane space, the endoplasmic reticulum, and the bacterial periplasm. In each system, disulfide bonds are introduced into substrate proteins and electrons are removed (oxidation of substrate proteins). A disulfide generating enzyme oxidizes a disulfide carrier that in turn oxidizes the substrate.
Mitochondrial precursor proteins are synthesized in the cytosol and subsequently imported into mitochondria. The import of mitochondrial intermembrane space proteins is coupled with their oxidative folding and governed by the mitochondrial intermembrane space import and assembly (MIA) pathway. The cytosolic steps that precede mitochondrial import are not well understood. We identified a role for the ubiquitin-proteasome system in the biogenesis of intermembrane space proteins. Interestingly, the function of the ubiquitin-proteasome system is not restricted to conditions of mitochondrial protein import failure. The ubiquitin-proteasome system persistently removes a fraction of intermembrane space proteins under physiological conditions, acting as a negative regulator in the biogenesis of this class of proteins. Thus, the ubiquitin-proteasome system plays an important role in determining the levels of proteins targeted to the intermembrane space of mitochondria. http://mcb.asm.org/
Mitochondria contain several hundreds of proteins. The mitochondrial content is regulated by the uptake and degradation of proteins. Stabilization of protein structure by disulfide bonds was proposed to drive protein accumulation in the intermembrane space of mitochondria. However, it remained unknown if structural alterations could lead to protein escape through the physiological barrier formed by the outer mitochondrial membrane. In this work, we present evidence for size-dependent retrograde movement of mitochondrial proteins to the cytosol. We identify the translocase of the outer mitochondrial membrane channel protein Tom40 as an exit route. Our results indicate that the retro-translocation serves as an important surveillance mechanism that regulates the abundance of intermembrane space proteins in response to changes in cellular physiology.
The content of mitochondrial proteome is maintained through two highly dynamic processes, the influx of newly synthesized proteins from the cytosol and the protein degradation. Mitochondrial proteins are targeted to the intermembrane space by the mitochondrial intermembrane space assembly pathway that couples their import and oxidative folding. The folding trap was proposed to be a driving mechanism for the mitochondrial accumulation of these proteins. Whether the reverse movement of unfolded proteins to the cytosol occurs across the intact outer membrane is unknown. We found that reduced, conformationally destabilized proteins are released from mitochondria in a size-limited manner. We identified the general import pore protein Tom40 as an escape gate. We propose that the mitochondrial proteome is not only regulated by the import and degradation of proteins but also by their retro-translocation to the external cytosolic location. Thus, protein release is a mechanism that contributes to the mitochondrial proteome surveillance. http://www.pnas.org/

Protein transport across outer and inner mitochondrial membranes; Protein import machinery; Protein Import Pathways of Mitochondria Most mitochondrial proteins are synthesized as precursors in the cytosol and are imported by the translocase of the outer mitochondrial membrane (TOM complex). (A) Presequence-carrying (cleavable) preproteins are transferred from TOM to the presequence translocase of the inner membrane (TIM23 complex), which is driven by the membrane potential (Dc). The proteins either are inserted into the inner membrane (IM) or are translocated into the matrix with the help of the presequence translocase-associated motor (PAM). The presequence are typically cleaved off by the mitochondrial processing peptidase (MPP). (B) The noncleavable precursors of hydrophobic metabolite carriers are bound to molecular chaperones in the cytosol and transferred to the receptor Tom70. After translocation through the TOM channel, the precursors bind to small TIM chaperones in the intermembrane space and are membrane inserted by the Dc-dependent carrier translocase of the inner membrane (TIM22 complex). (C) Cysteine-rich proteins destined for the intermembrane space (IMS) are translocated through the TOM channel in a reduced conformation and imported by the mitochondrial IMS import and assembly (MIA) machinery. Mia40 functions as precursor receptor and oxidoreductase in the IMS, promoting the insertion of disulfide bonds into the imported proteins. The sulfhydryl oxidase Erv1 reoxidizes Mia40 for further rounds of oxidative protein import and folding. (D) The precursors of outer membrane b-barrel proteins are imported by the TOM complex and small TIM chaperones and are inserted into the outer membrane by the sorting and assembly machinery (SAM complex). (E) Outer membrane (OM) proteins with a-helical transmembrane segments are inserted into the membrane by import pathways that have only been partially characterized. Shown is an import pathway via the mitochondrial import (MIM) complex. www.cell.com
https://www.biochemie.uni-freiburg.de
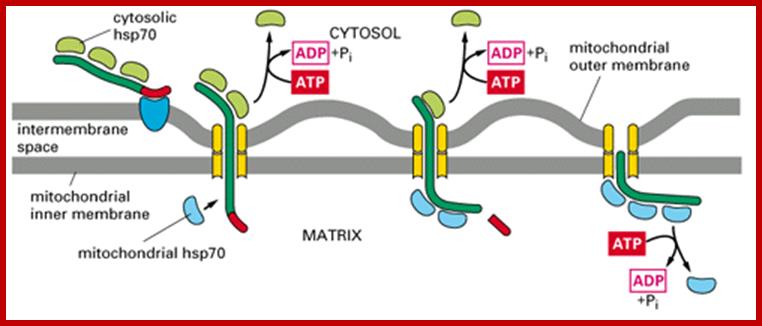
Most of the mitochondrial proteins are synthesized as precursor with cleavable N-end cleavable sequences. The cleavable sequences for import are charged amphipatheic alpha helices and they are recognized mitochondrial membrane bound receptors on the outer membrane (TOM). After translocation into perimitochondrial space the preproteins transferred to inner membrane translocator protein complexes (TIM23). The presequence associated motor protein (PAM) drives the preproteins are translocated into mitochondrial matrix. Once translocated the presequences are cleaved in the matrix. Some cleavable proteins get arrested by TIM23 and later they are released into inner membrane. In some proteins are translocated into matrix and followed by insertion into inner membrane matrix
www.cc.scu.edu.cn; cc.scu.edu.cn
www.cc.scu.edu.cn; cc.scu.edu.cn
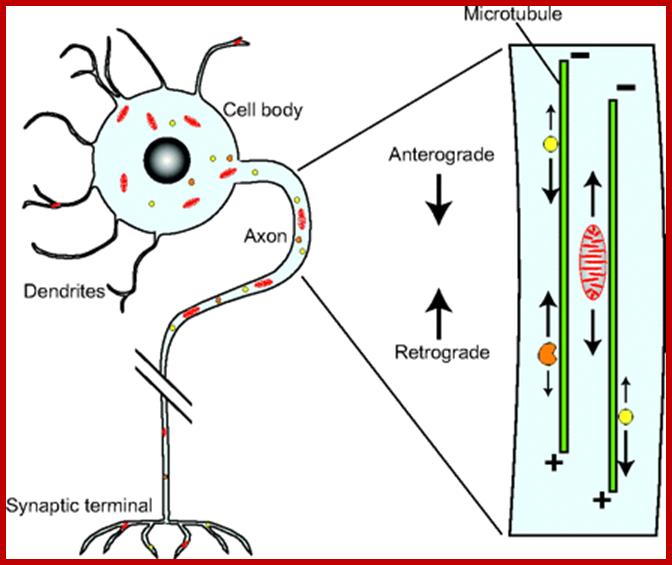
Movement of Mitochondrai:
Mitochondrial movement in a cell, where some cells are long reaching several micrometers from the cell body ex. Axons of nerve cells. Most of the movements are anterograde example from the cell base towards the synaptic surface of the neuronal cell and the backward movement is called retrograde. Anterograde movement is from the base or cell body of the cells to toward microtubule plus end and they use Kinesin motor proteins. Retrograde transport is from plus end of MT to negative end or synaptic surface to basal cell body. Similarly many other cellular components are transported forward and backward.
http://jcs.biologists.org/
http://bio100.class.uic.edu/
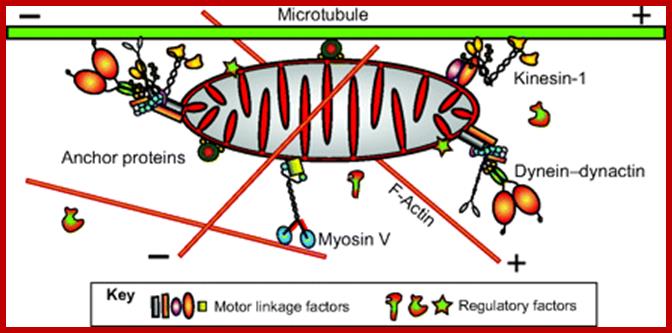
Important structures involved are microtubules which use motor proteins Kinesins forward (anterograde) to microtubule (MT) plus ends and Dyneins are used to transport from plus ends towards minus ends called retrograde movement. Mitochondria are also transported in axon by the same process. New mitochondria generated in cell bodies are transported to the nerve ends.
http://jcs.biologists.org/
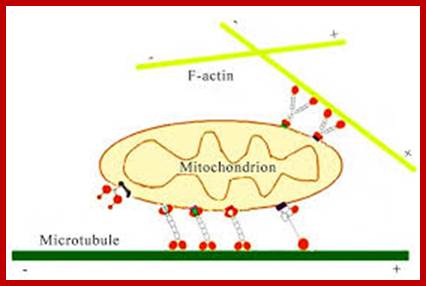
Axonal transport from neuron cell body to terminal region by retrograde movement of endosomes all along the length of microtubule from minus end toward plus ends. Actins are also involved in mitochondrial transport using myosin. https://publish.illinois.edu; http://file script.org
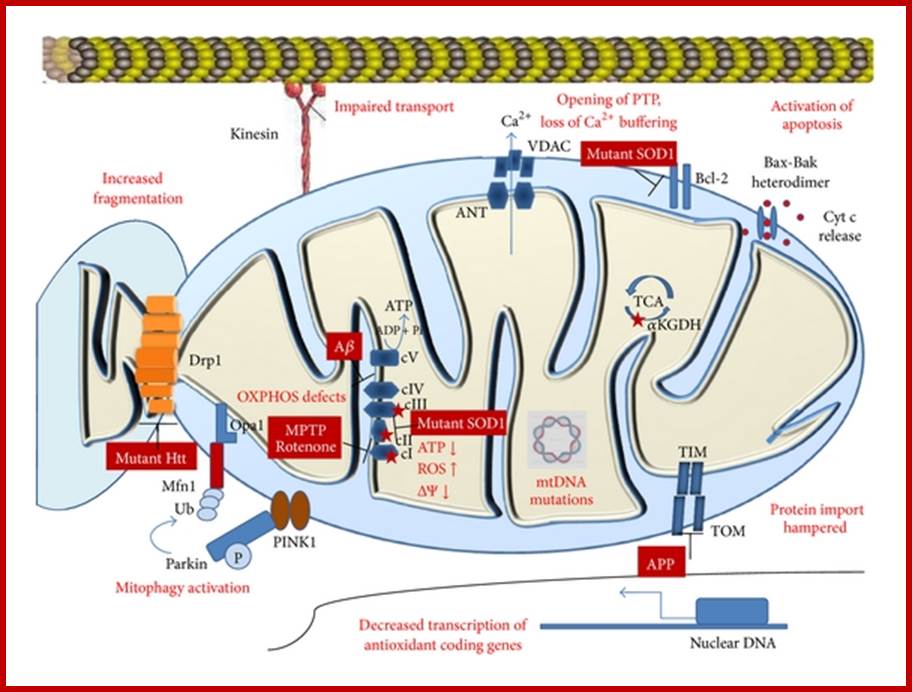
Mitochondrial changes in neuro-degeneration; Most neurodegenerative diseases result in defects in oxidative phosphorylation, respiratory dysfunction, increased ROS production, lowered mitochondrial membrane potential, decrease in synthesis of antioxidants, mtDNA mutations, impaired protein import, increased fragmentation of mitochondria, and activation of mitophagy and apoptosis. Sources of reactive oxygen species are marked by red stars. Abbreviations: Htt, Huntingtin; Ub, ubiquitin; ΔΨ, membrane potential; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; ROS, reactive oxygen species; mtDNA, mitochondrial DNA; Aβ, amyloid beta; APP, amyloid precursor protein; TIM, translocase of inner membrane; TOM, translocase of outer membrane; SOD1, superoxide dismutase 1; VDAC, voltage dependent anion channel; ANT, adenine nucleotide translocator; Pink1, PTEN-induced putative kinase 1; αKGDH, alpha ketoglutarate dehydrogenase; cyt c, cytochrome c.;ww.hindawi.com.
For More Information Read-www.grkraj.org
Uniparentall Inheritance of mitochondria in various groups of plant and animal systems:
Uniparental inheritance of mtDNA is observed in many sexually reproducing species. However, it may be accomplished by various different strategies in different species. Possible mechanisms are listed as below: 1)decrease in the content of mtDNA during spermatogenesis,2) elimination of mtDNA from mature spermatozoa, 3) prevention of sperm mitochondria from entering the oocyte, 4) active degradation of the paternal mtDNA in the zygote, 5) selective degradation of the whole paternal mitochondria in the zygote.
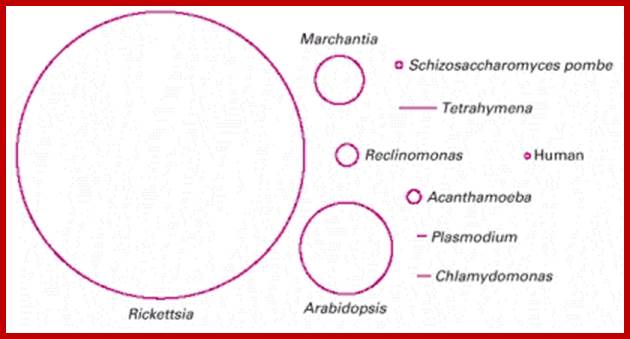
Size of mitochondrial DNA varies; http://www.sciencedirect.com/
Fate of Mitochondria found in Spermatazoids:
Mitochondria are inherited by mother and not by father; it is always maternal inheritance. Any sperm that enters fertilized egg, the sperm mitochondria is degraded due to a tag that is carried during spermatogenesis. However mitochondrial population is same in all for it lacks proper repair mechanism and proof reading capabilities. Mutation rate of mitochondrial DNA is ten times higher than nuclear DNA. More than 30 ubiquitinating enzymes have been identified as important regulators of spermatogenesis.
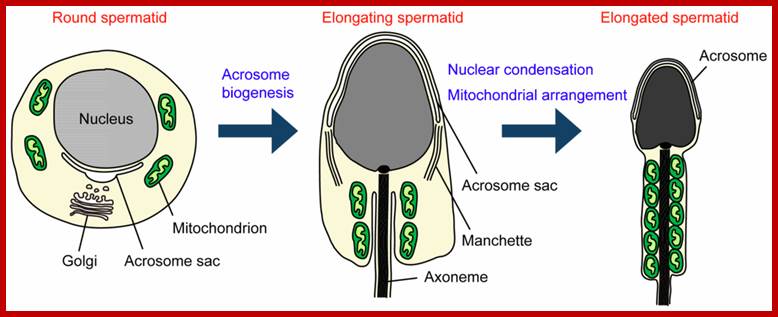
Morphological changes during spermiogenesis. In elongated spermatozoids mitochondria are aligned along the anterior part of the flagellum and tightly packed to form the helically arranged mitochondrial sheath; http://www.mdpi.com/
Mature sperm cell is slender; in the middle part , the mitochondria are thick and rig shaped. The DNA of the nucleus is maximally condensed; Mitochondria present in mid piece get destroyed by using ubiquitinated surfaces by proteasomes,
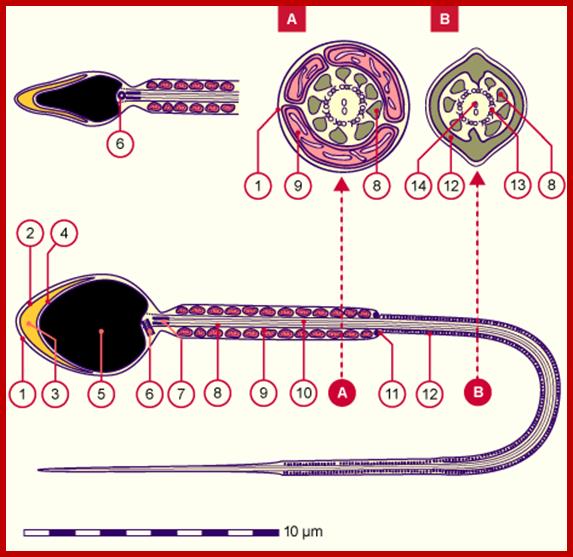
Neck contains two centrioles, Mid-piece consists of a sheath of ring-shaped mitochondrial grouped, the principal piece has a sheath of a ring of fibers around axoneme, The tail consists of only the 9+2 structure of the axoneme, The mature sperm cell is approximately 60um long and completely enveloped by the plasma membrane.
1.Plasma membrane, 2. Outer acrosomal membrane, 3.Acrosome, 4. Iner acrosomal membrane, 5.Nucleus, 6. Proximal centriole, 7. Rest of the distal centriole, 8. Thick longitudinal fibers, 9. Mitochondria, 10. Axoneme, 11. Annulus, 12. Ring fibers; A.Head, B. Neck, D. Principal Piece, and E.End piece. http://www.embryology.ch/

http://leavingbio.net/
Maternal inheritance of mitochondrial DNA has long been regarded as a major paradox in developmental biology. While some confusion may still persist in popular science, research data clearly document that the paternal sperm-borne mitochondria of most mammalian species enter the ooplasm at fertilization and are specifically targeted for degradation by the resident ubiquitin system. Ubiquitin is a proteolytic chaperone that forms covalently linked polyubiquitin chains on the targeted proteinaceous substrates. The polyubiquitin tag redirects the substrate proteins to a 26-S proteasome, a multi-subunit proteolytic organelle. Thus, specific proteasomal inhibitors reversibly block sperm mitochondrial degradation in ooplasm. Lysosomal degradation and the activity of membrane-lipoperoxidating enzyme 15-lipoxygenase (15-LOX) may also contribute to sperm mitochondrial degradation in the ooplasm, but probably is not crucial. Prohibitin, the major protein of the inner mitochondrial membrane, appears to be ubiquitinated in the sperm mitochondria. Occasional occurrence of paternal inheritance of mtDNA has been suggested in mammals including humans. While most such evidence has been widely disputed, it warrants further examination. Of particular concern is the documented heteroplasmy, i.e. mixed mtDNA inheritance after ooplasmic transplantation. Intracytoplasmic sperm injection (ICSI) has inherent potential for delaying the degradation of sperm mitochondria. However, paternal mtDNA inheritance after ICSI has not been documented so far. http://www.ncbi.nlm.nih.gov/
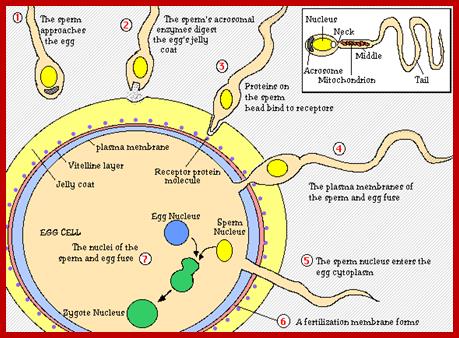
http://image.slidesharecdn.com/
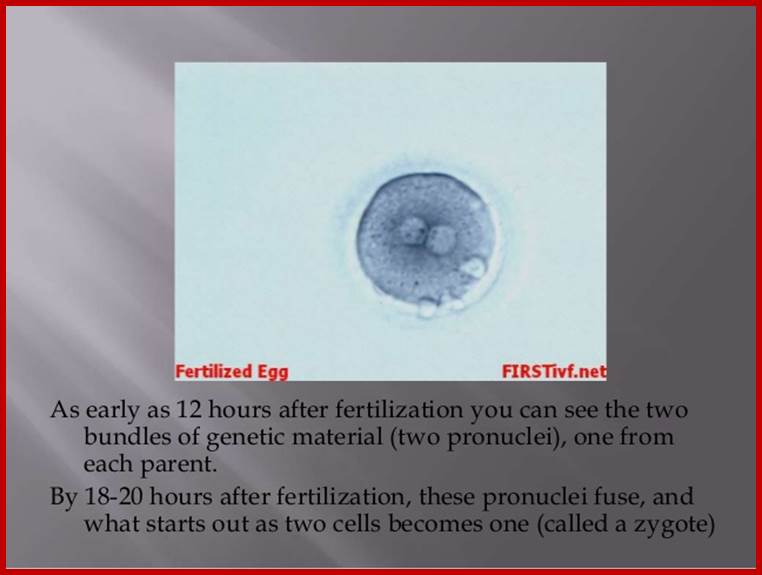
http://image.slidesharecdn.com/fertilization notes
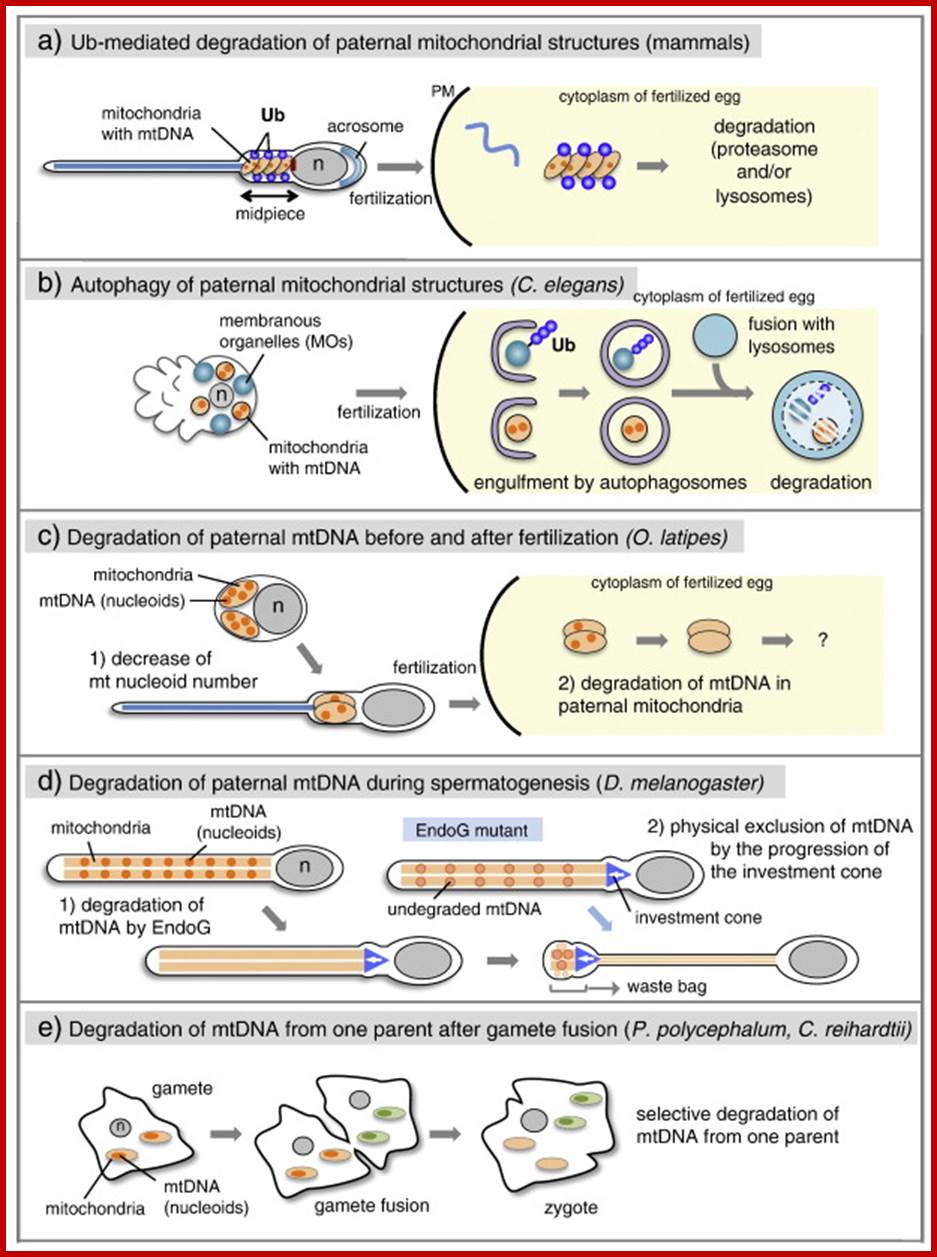
Multiple mechanisms of paternal mtDNA elimination.
In mammals, paternal mitochondria are modified by ubiquitin binding to specific proteins on mitochondria during spermatogenesis and degraded by the proteasomes and/or lysosomes after fertilization. (b) In Caenorhabditis elegans, paternal mitochondria and membranous organelles (MOs) are engulfed by autophagosomes and targeted for lysosomal degradation after fertilization. Ubiquitination is detectable on MOs. (c) In Oryzias latipes, the number of mtDNA nucleoids decreases during spermatogenesis. After fertilization, paternal mtDNA is further degraded before the destruction of the mitochondrial structure. (d) In Drosophila melanogaster, paternal mtDNA is degraded by EndoG during spermatogenesis. In EndoG mutants, the remaining mtDNA nucleoids are eliminated by investment cones and deposited into a waste bag. (e) In the isogamous species of Physarum polycephalum and Chlamydomonas reinhardtii, mtDNA from 1 parent is selectively degraded in the mitochondria after gamete fusion.
Uniparental inheritance of mtDNA, as well as chloroplast DNA (cpDNA) or plastid DNA (ptDNA), is also observed in plants ... However, in most mammals, including humans and mice, the paternal mitochondria and their mtDNA do enter the oocyte cytoplasm upon fertilization; In mammals paternal mitochondria are ubiquitin mediated degradation takes place in the fertilized egg.
cpDNA is maternally inherited in the four-o'clock plant (Mirabilis jalapa) . In Sequoia sempervirens, in which mtDNA and cpDNA are paternally inherited. n the geranium plant (Pelargonium zonale), cpDNA is inherited maternally, paternally or biparentally . When zygotes receive cpDNA from both parents, cpDNAs from each parent are rapidly segregated following cell division, resulting in the mature plant inheriting clonal sectors of cells that are homoplastic for only 1 type of cpDNA. In angiosperm species, such as Arabidopsis thaliana, both mtDNA and ptDNA are maternally inherited and . In A. thaliana, the active degradation of paternal mtDNA and ptDNA takes place during pollen development.
The content of mtDNA and cpDNA gradually decreases during meiosis, and these DNAs are no longer detectable in mature pollen. The wild-type DPD1 encodes a pollen-specific Mg2 +-dependent exonuclease, which is transported into both chloroplasts and mitochondria, suggesting a direct role for this exonuclease in the degradation of mtDNA and ptDNA. http://www.sciencedirect.com/; http://biology-themiracleoflife.blogspot.in/; http://www.ncbi.nlm.nih.gov/