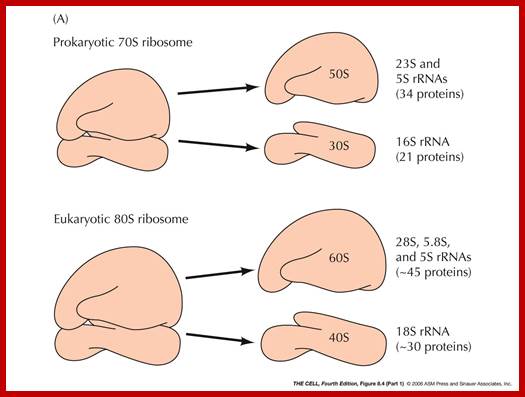
Ribosomes:
Introduction:
Ribosomes are made up of rRNAs and proteins for they act as structural components of Ribosome organelle. The ribosome in its entirety is constructed on ribosomal RNA as a scaffold on which riboproteins are sequentially built to produce a highly dynamic structure, which has astounding abilities to function as translation machine.
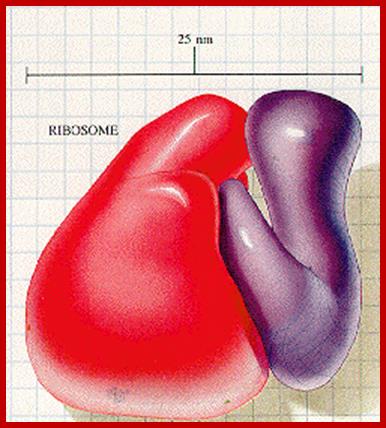
An excellent over view of ribosomal subunits hugging to each other.www.library-online.blogspot.com; http://urei.bio.uci.edu/;https://sites.google.com
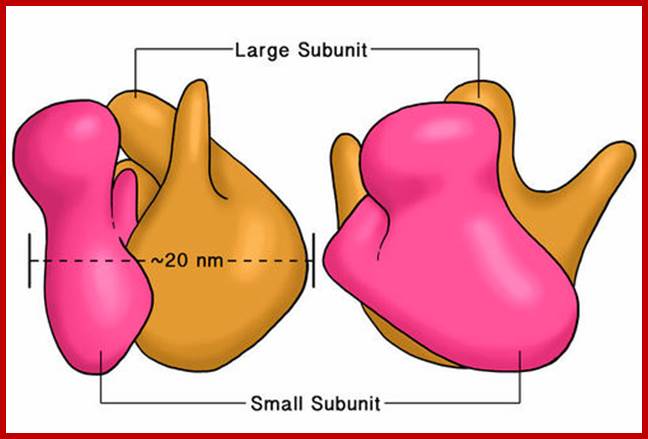
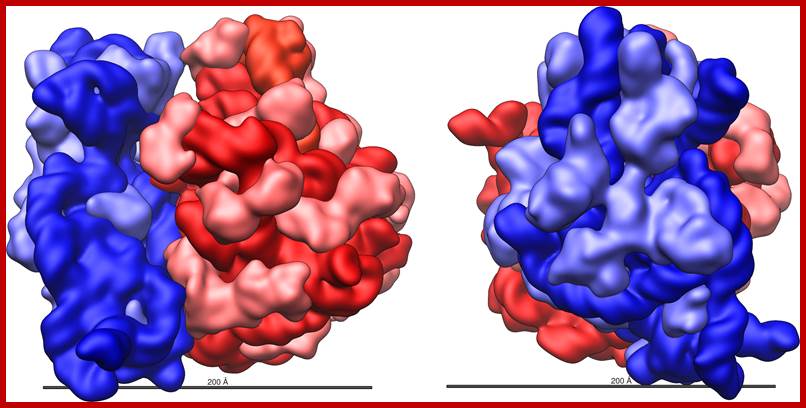
Three-dimensional views of the ribosome, showing rRNA in dark blue (small subunit) and dark red (large subunit). Lighter colors represent ribosomal proteins.;www.scienceprofonline.com, en.wikipedia.org; www.biology.about.com
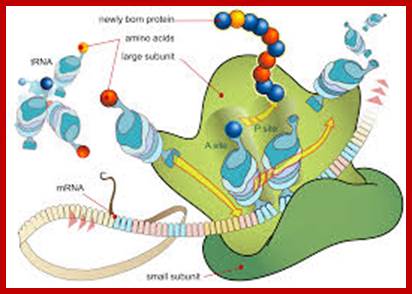
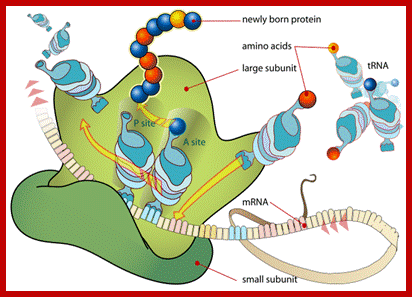
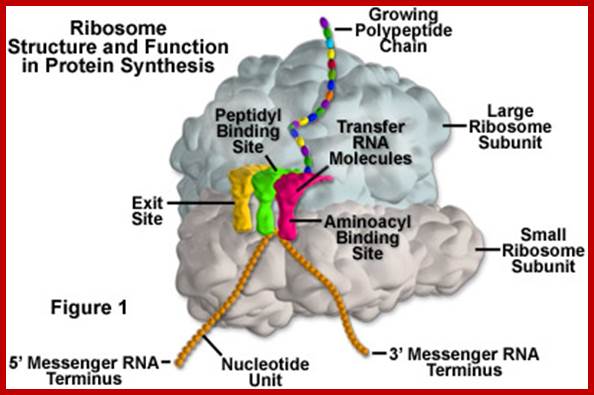
Diagram showing the translation of mRNA and the synthesis of proteins by a ribosome;www.princeton.edu; chemistry.about.com
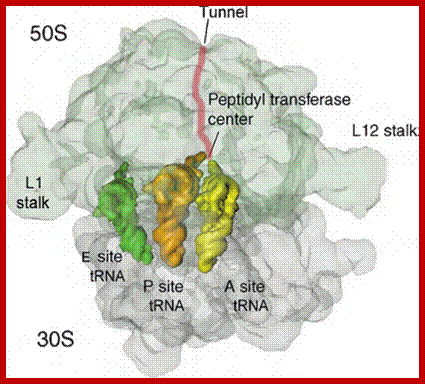
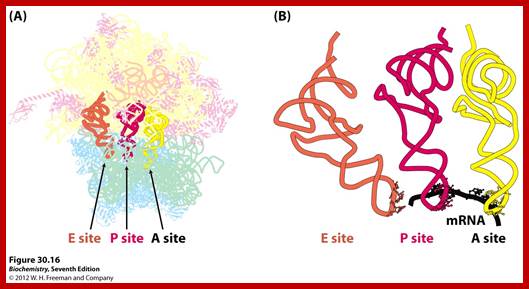
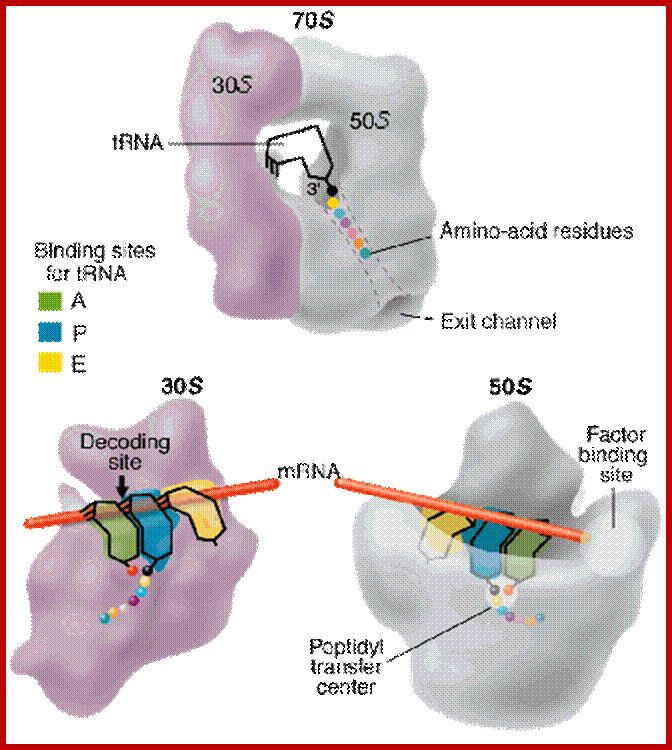
Ribosome binds to both mRNA and tRNA; structural features of tRNAs at different positions is shown; Binding of tRNA during translation, binding positions such as E, P and A sites are shown. www.oregonstate.edu;www.chegg.com

http://oregonstate.edu/
Distribution:
Ribosomes are found in almost all organisms except viruses. An E.coli cell may contain 15000 to 20000 ribosomes at any given time, but an active eukaryotic cell may have 10-20 times the number of prokaryotic cells, perhaps in millions. Oocytes of certain amphibians’ posses’ three million ribosomes per cell and the same is stored for the future use. While in prokaryotes, ribosomes are distributed through out the cell, eukaryotic cells contain different classes of ribosomes and they are located in different loci like cytoplasm, mitochondria and plastids. Cytoplasmic 80s ribosomes are either bound to endoplasmic membrane or free. The majority of the so-called free ribosomes are found located in the intersection of microtrabacular (?) and actin filament network. On the contrary cellular organelles like chloroplast and mitochondria themselves contain another class called 70s ribosomes, which are more or less similar to that of bacterial ribosomes. In the Oocytes of chicks and lizards, ribosomes are aggregated on membranes into crystalline structures. They remain inactive till they are required at some stage of development.
Class of ribosomes:
Ribosomes can be isolated by magnesium precipitation. If some ribosomes, obtained from a eukaryotic organism, are subjected to density gradient ultracentrifugation, ribosomes settle into two distinct bands. Based on the sedimentation values, determined by Svedberg, they can be distinguished into 70s and 80s ribosomes. The 80s ribosomes are found in cytoplasm, whereas 70s types are found in mitochondria and chloroplasts. The 70s type are smaller and 80s are little larger. However, prokaryotes contain only one kind of ribosomes i.e. 70 type. The 80s and 70s ribosomes can be further distinguished by their sensitivity to chloramphenicol (CAP) and cycloheximide (CHI) respectively. The 70s ribosomal mediated protein synthesis is inhibited by chloramphenicol, while 80s ribosomal protein synthesis is inhibited by CHI.
Chemical composition:
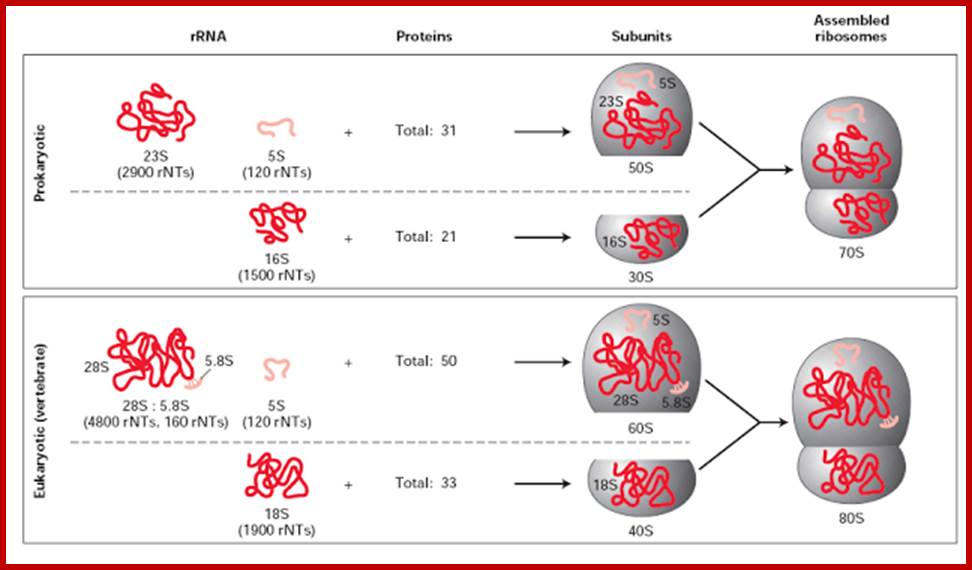
Components of Ribosomes:
|
Types |
RNA size |
Number of proteins |
Methylations |
Functions |
|
70 S ribosomes |
Coded by several genes |
|
|
30 or more methylations |
|
30s subunits |
16s RNA, 1540-42 ntds |
21 (s1 to s21) |
10 at 2’OH, 2,methyl adenines, 2,dimethyl guanines |
Help in processing and folding |
|
50S subunits |
23s RNA, 2900 ntds; 5s RNA, 120 ntds |
31, L1 to L31 |
20 at 2’OH of sugars |
|
|
80S ribosomes: |
Coded by hundreds of genes, rRNA genes located on chromosomes 12,13,14,21 and 22 (in humans) |
|
|
>100 sites for methylations and 100 sites for pseudouridenylations Yeast has 43 pseudo uridines |
|
40S subunits |
18s RNA;( 1843 Or 1900 ntds) |
33; S1 to s34 |
43 to 44 methylations at 2’OH groups, plus conversion of Uridine into pseudo-Uridines |
|
|
60s subunits |
28s-RNA;(4718- 4800 ntds); 5.8s RNA;(160ntds); 5s RNA;(120ntds);
|
49; L1 to L45-50 |
74 methylations at 2’OH of sugars, Methylation at adenine, Methylation at guanine, plus conversion of Uridine into pseudo-Uridines |
|
|
Mitochondrial ribosomes: 70s like (general); Fungus-73s; Maize-78s; |
16s 12s |
-1560 ntds, 48 proteins -29 proteins
|
|
|
|
Chloroplast ribosomes: 70s (50s and 30s) |
23s RNA 16s RNA 4.5s RNA? |
23sRNA, 5s RNA, 4.5s, 16s RNA |
|
|
|
|
|
|
|
|
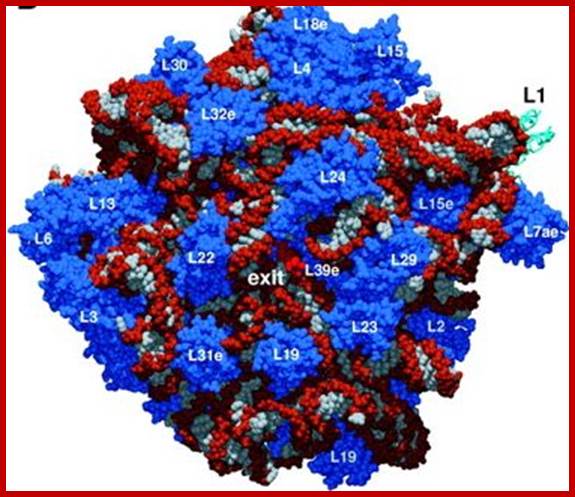
Prokaryotic Ribosomal RNA and Riboproteins:

This figure shows 70S ribosomal subunits
A simple diagram showing subunit components of ribosomal subunits www.is.muni.cz
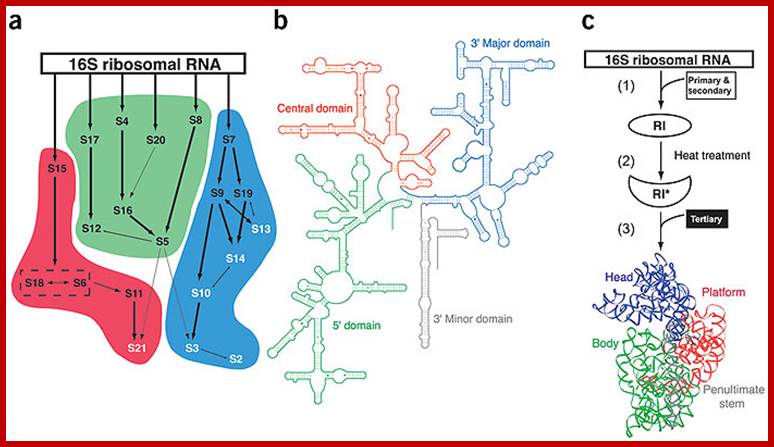
In vitro assembly of 30s ribosomes showing rRNA; Primary and secondary ribosomal proteins in black and tertiary proteins in white. S6 and S18 heterodimers are enclosed in box; b. secondary structures of 16S rRNA one can observe four domains 5’ central , 3’ major and 3’ minor domains are in green, red and gray; C. schemating representation of in vitro 30S ribosomal assembly. Kristi L Holmes & Gloria M Culver; http://www.nature.com/
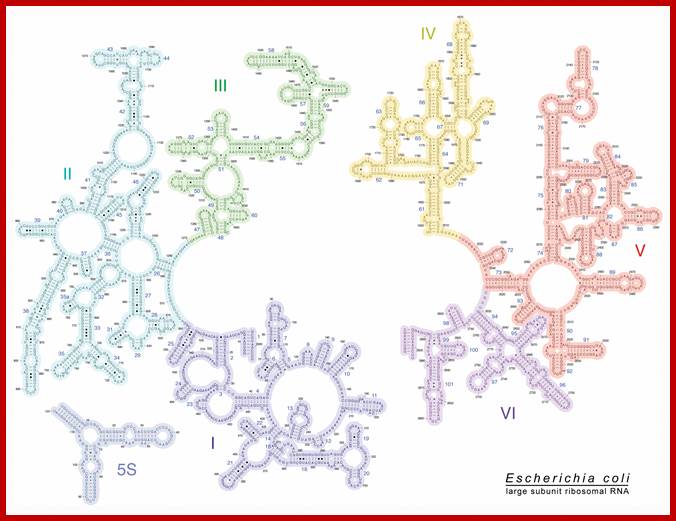
E.coli 23S rRNA secondary structural features; http://rna.ucsc.edu/
![]()
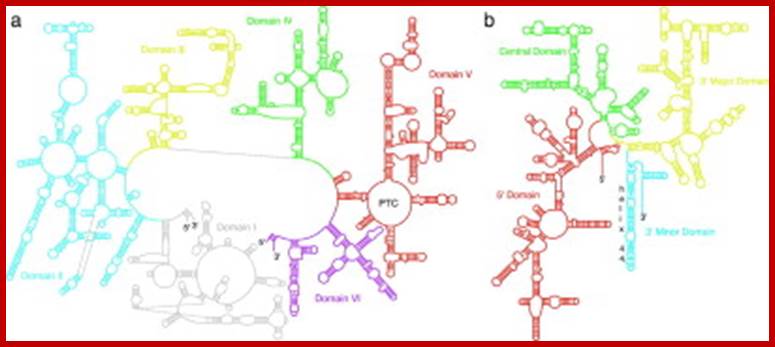
Secondary Structure Diagrams of the 23S and 16S rRNAs: The different domains are color coded. (a) The 23S rRNA of H. marismortui. (b) The 16S rRNA of T. thermophilus. Locations of helix 44 and the peptidyl transferase center (PTC) are indicated. Diagrams were modified with permission of R. Gutell (http://www.rna.icmb.utexas.edu)
- Secondary structures of each of the rRNAs have been determined by their sequence analysis.
- The 16s rRNA and 23s rRNA, each of them, show four domains and each of them are distinguished by the binding of specific riboproteins.
- The 16s RNA’s domain I starts from 5’ end progresses into domain II, III and IV in an order.
- At the 3’ end of the IV th domain of the 16s rRNA it has a small segment with a sequence that binds to the 5’ end of non coding Shine-Delgarno sequence which is a leader sequence of mRNAs.
- The sequence is 3’ AUUCCUCCACUAG—5’.
- Similarly there are specific ribo-protein binding sites in each of the domains, ex. S4 & s20 bind to domain I, s8 7 & s15 bind to domain II, s7, 9,13 and 19 bind to domain III; thus each of the binding domain can be identified.
- Binding of tRNA and other factors to specific regions have been discerned by a variety of techniques such as electron microscopy, immuno labeling, neutron scattering techniques.
- Even eukaryotic subunit rRNAs show such domains identified by their ability to bind to certain riboproteins and other RNA species such as tRNA and other translational factors.
- Some commonality of the sequence and secondary structures can be observed when one compares the 5’end of the 23s RNA of prokaryotes with that of 5’ ends of eukaryotic 5.8sRNA. A secondary structure of rRNAs suggests the overall structural features of ribosomes.
- It can be discerned that rRNAs from eukaryotes also show similar structural features.
Riboproteins (Prokaryotic):
- Riboproteins, isolated from small and large subunits, have been numbered as small and large riboproteins.
- They are numbered as s1 to s21 and L1 to L31 respectively. The numbering is based on the mobility of each of the riboprotein subunits on a 2-D polyacrylamide gel. The S1 is the largest protein found at the left top most corner of the gel and s21 is the smallest, found at the right bottom most corner of the gel.
- Each of the riboproteins from both 70s and 80s ribosomes have been purified and antibodies have been obtained and many proteins have been subjected to X-ray diffraction, immuno-diagramming, neutron scattering, electron microscopy, NMR and cross linking to other riboproteins
- In prokaryotes s20 and L26 are common to both ribosomal subunits and located at the interface of the subunits.
- L7 and L12 are found 4 copies each. Their N- terminal is acetylated and forms stable complex with L10.
- But most of the riboproteins are made in single copies.
- Amino acid sequence of most of the riboproteins has been determined, where the number of amino acids ranges from 46 (of smallest subunit at right bottom most in the gel) to 557 in the largest subunit s1 (Left top most in the gel).
- L1 is the largest with 25 KD and L34 with 45KD.
- There is sequence homology among the protein subunits, but in general, they are rich in Lysine and Arginine, both are basic amino acids.
- Structural motifs of riboproteins show 3 b sheets and 2 a helical strands and resemble that of Sn RNPS, rho, and eIf-4B. Such motifs are called RRMs (rRNA Recognition Motifs).
- Most of the riboprotein genes have been identified and their operons have been established.
- Each operon consists of several genes controlled by a single promoter element.
- In prokaryotes the riboprotein genes have been organized into seven clusters called rRNA operons.
- Some of these operons not only contain riboprotein genes but also contain other genes like RNAP subunits and translational factors.
- Their synthesis is regulated at translational level, which depends upon the availability and the abundance of rRNA segments.
- If the concentration of rRNA falls short, the translation of their mRNAs is inhibited by their own proteins by binding to specific regions of the mRNAs they prevent translation, which is called autogenous regulation.
- Subunits of riboproteins act as self-assembling systems with rRNA providing a template or threading for sequential assembly.
- As rRNA is synthesized (synthesized as a precursor RNA), it is methylated at specific sites, which acts as recognition points for processing RNA and also assembly of proteins.
- Assembly of riboprotein with rRNA is hierarchical, in the sense, some proteins bind first to certain RNA sequence derived secondary structures, then another set of proteins bind either by RNA- protein interaction or protein-protein interaction or by both.
- There is a gradual build up of the assembly line, which ultimately produces a highly organized and a dynamic structure, where the position of each protein is fixed with respect to each RNA fold or sequence.
- Though it exhibits a specific 3-D shape, it is not a rigid structure but it always undergoes conformational changes according to the binding of other translational components or regulatory factors. Structurally, as well as functionally, ribosomes are flexible and highly dynamic.
- In the overall organization of ribosomal structure, some regions don’t have any RNA but only proteins. The whole structure appears as porous; through some of RNA folds protrude to the surface.
Assembly:
- Assembly or association of riboproteins with rRNA is sequential and stepwise.
- Methylation of 2’OH of ribose sugars and at adenine and guanine nucleotides at specific position is critical, and such methylations are performed by specific methylases and they use sequence driven secondary structures as motifs for identification of sites.
Assembly of small ribosome subunits:
16sRNA + 16 s riboproteins à 21 s particles (can assemble at 20^oC),
21s particles + 6s riboproteins >à 26 s particles,
26 s particles ----> 30 s particles.
Assembly of Large Ribosome subunits:
23SRNA + 5sRNA -à 33 s particle,
33 s [articles -à 41 s particles,
41 s particles -à 50s particles
Next, let’s put red circles around the ribosomal proteins for which there is experimental evidence to support a moonlighting role (the moonlighting roles are listed below the
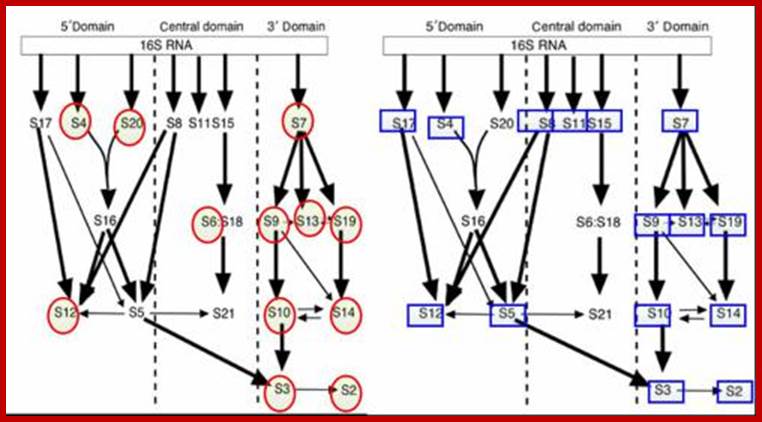
http://designmatrix.wordpress.com/
During dissociation also, certain subunits dissociates fast, even at the earliest steps of preparation; they are called split proteins. Such proteins are found both in small and large subunits. Even during assembly, certain proteins associate at 0^oC, this is because great affinity of some proteins to certain RNA sequence. Cold sensitive mutants block such assembly; they are called Subunit Assembly Defective mutants (SAD mutants). Proteins, which associate, first are hard to disassociate and they are called core groups, and proteins, which assemble last, are the first to dissociate. The following figure depicts sequential steps in the assembly.
rRNA 5’--------------------------------------------------------------------------3’
I I I I
1st level I s4 I I s8
2nd level s15 I s20 s7
3rd level s17 s13
4th level s16
5th level s12 s9 s19
6th level s18 s5
Assembly sequence:
30s = 17.5sRNAàs4,s8,s15-às1,s5,s7,s13--->s2,s3,s6,s9,s10
s17, s20 s16, s21 s11, s12, s14, s18/19
50s= 25sRNA--->L1,4,5,8,9,10---->L3,7,11,14-->L2, 6,12,10,28,31,32,
13,17,18,20, 15, 19, 23
21,21,22,23,
24,25,27,29,
30, 33.
30s [16s RNA] O^oC 40^oC O^oC
+[ s21 proteins]--------------------> 21s--------------->26s------------->30s
50s [23sRNA] o^oC 44^oC O^oC 50^oC
+5sRNA+34L] ---------------->33s---------------->41s------------->48s----------->50s
Proteins]
- As the 5’ end of the precursor rRNA emerges during its synthesis, s4, s8 and s15 bind to this region tightly. Then s17 and s7 join directly on to the RNA, later other proteins join by protein-protein or protein-RNA interactions.
- Proteins s1, 3, 4,5,9,12,18 and the 3’ end of 16s RNA are involved in mRNA binding.
- Peptidyl transferase function at P-site involve proteins L-2, 11, 15,16,18,23, and 27 in association with 23s RNA.
- About 40 ntds long region of 16S RNA is located in the platform of 30s ribosomal subunit.
- Peptidyl transferase occupies valley in the ribosome.
- Two L7 and two L12 together act as GTPase.
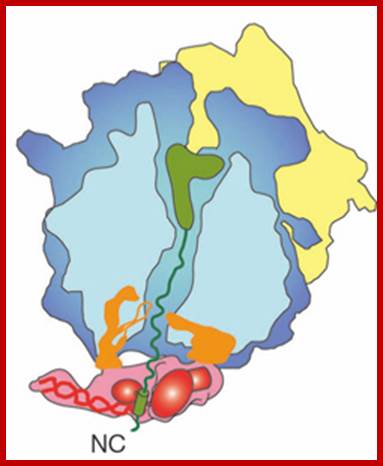
The large subunit of the ribosome is in blue, the small subunit in yellow. The canal in the large subunit is where the newly synthesized peptide (protein) is pushed through (think of it as the birth canal of the ribosome. As new amino acids are being added to the top where the green blob is (the green blob is a tRNA – I will not explain it here any further) the newly synthesized peptide (in green) is being pushed out, or down in this diagram, towards the exit. At the exit site, SRP (here in red) sits and in this diagram is holding the part of the polypeptide that encodes the signal sequence (the green cylinder). SRP is also making contacts to two subunits of the ribosome (the two orange blobs). When bound to the signal sequence SRP makes a total of four contacts to the ribosome. Some of these contacts are shared with another ribosome cofactor, trigger factor (TF), which acts as a chaperone for the newly emerging nascent chain. Since TF and SRP share binding sites they may be mutually exclusive. In fact when SRP holds on to a newly made signal sequence and engages the SRP receptor in the ER, TF is known to be released from the ribosome. http://scienceblogs.com/
The ribbon diagram shows the positioning of tRNA on large ribosomal surface; A, P and E sites; http://liberary-online.blogspot.com
Role of rRNA in protein synthesis (Prokaryotic):
- In the molecular organization of ribosomes, both RNA and proteins are ordered and occupy certain specific invariant positions and perform specific function. As a 3-D structure, it goes through several conformation changes with each binding events and catalysis.
- The 3’ terminus 16s RNA of 30s ribosome directly interacts with 5’ end Shine-Delgarno sequence of mRNA and facilitates initial binding of it ribosomal surface so as to bring the first codon exactly to P site.
- Specific regions of 16s RNA interact with tRNA for the binding at P and A sites.
- 23 S RNA interacts with CCA terminus of peptidyl tRNA.
- It is envisioned that there is RNA-RNA interaction and protein –protein interaction as well as protein-RNA interaction, thus both subunits are held together while they perform functions.
- There must be some proteins, which perhaps act as motor proteins in moving ribosomes on single stranded mRNA in ATP dependent manner, similar to helicase.
- Cleavage of 3’ region of 16S RNA by E3 Colicin abolishes initiation of translation.
- Methylation of Adenine at 6th position and at 3’ end (di methylation to Adenine) of 16s RNA, facilitates dimrization of 30s and 50s units. If methylases are absent dimrization fails.
- Sensitivity to Kusugamycin depends upon methylation or absence of methylation at the above-mentioned sites. Mutation at these sites abolishes sensitivity to the drug, but methylation at these sites makes it Kusugamycin resistant.
- Kusugamycin blocks initiation of translation by releasing F-met tRNA from ribosomal surface.
- Mutation in the of 3’ end of 16s RNA suppress terminator codon function.
- A mutation in rRNA can lead to frame shift function, because recognition between mRNA and rRNA doesn’t take place.
- A region at 1400th ntds in 30s, is directly involved in the binding of peptidyl tRNA at P-site.
- Both 16s and 23s RNAs are involved in organizing A and P site.
- The CCA-end of tRNA at P- site protects 23SRNA from RNase digestion.
- The 23s rRNA is involved in organizing the exit site E, found in 50s subunit.
- The 23s RNA is involved in catalyzing peptide bond formation; hitherto it is believed that an enzyme called di peptidyl transferase found in larger ribosomal subunit is involved in peptide bond formation.
Structural Features of Ribosomes (Prokaryotic):
Structurally prokaryotic ribosome has 200 x 220 A^o dimension and the size of eukaryotic ribosome is slightly larger.
- For initiating translation process, to begin with the small subunit should be separated from the large subunit; it is at later part of initiation both subunits join.
The larger subunit looks like a cup shaped palm having a central protuberance curved inwards, a blunt thumb like structure and a last finger like structure projecting outwards.
- The extended finger acts like a stalk and contains L7/L12 with GTPase activity.
The central protuberance contain 5s RNA.
- The valley is located between the blunt thumb like projection and central protuberance. The ridge or thumb like portion contains L1/9.
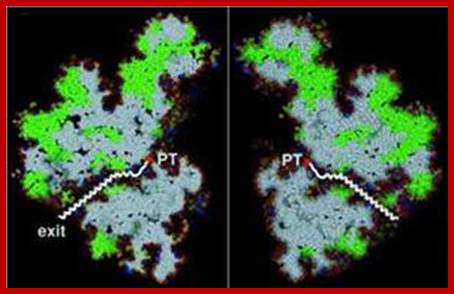
Actually the valley provides peptidyl transferase activity.
The large subunit has a narrow tunnel like region, which extends from peptidyl assembly site to exterior, through which nascent polypeptide chain is threaded through with NH3+ end ahead.
The length of the tunnel can hold about 25 to 30 amino acid long polypeptide chain and has the diameter to accommodate the chain.
It is at the posterior end, where polypeptide chain exits, contains a site for the binding of large ribosome to endoplasmic reticular membrane.
http://dwb.unl.edu/Teacher

This diagram shows a tunnel through which the nascent polypeptide threads through as it is translated. http://www.bioss.uni-freiburg.de/
The small subunit is split in the top region into a platform and a head; the space between them is called cleft.
- The 3’ end of the 16sRNA is located in the platform. It is through cleft region the mRNA is threaded and both P-site and A-site are located on the surface of platform and the base of the head.
Ribosomal site for the binding of mRNA to 16sRNA and the binding of initiation factors are located in the platform of 30s ribosome.
- Peptidyl tRNA and aminoacyl tRNA binding locations (P and A sites) are at the cleft region of 30S ribosome.
The small ribosomal subunit has an additional site called A site to the right of A site, where the incoming aa tRNAs are screened.
- The stalk (finger like) of 50s ribosome contain GTPase activity.
Peptidyl transferase activity is located in the valley of large unit.
- Large subunit also has sites for the binding of peptidyl (P) and aminoacyl tRNAs (A) in line with small subunit, plus it has another site for the exit (E) of decoded tRNA.
The tunnel is 100-120 A^o long, 25A^o broad and can hold approximately 20-30 amino acid long polypeptide chain.
- ER Membrane binding domain in the large subunit is by the side of tunnel exit.
Eukaryotic Ribosomes:
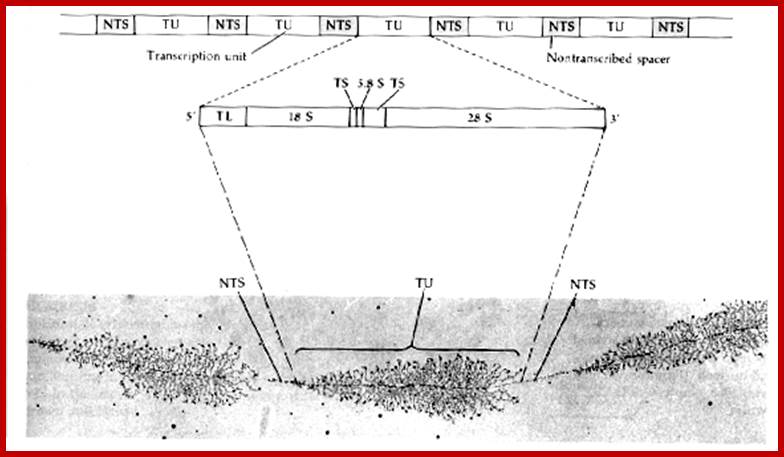
http://10e.devbio.com/
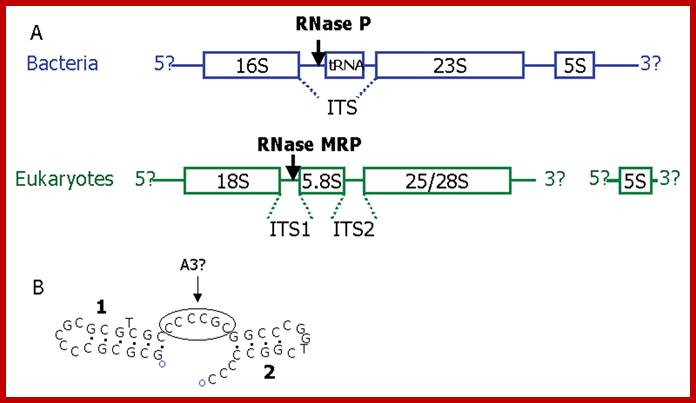
Promoter regions of Human MRP, P and U6.A. Organization of pre-rRNA based on [37]. In bacteria rRNA genes are co-transcribed as a polycistronic precursor (although exceptions are common). Most eukaryotes vary only in the length of their ITS regions, an extreme case being the microsporidian Encephalitozoon cuniculi which has completely lost its ITS2 having a fused 5.8S/28S subunit. RNase P and RNase MRP do not cleave the main transcripts but trim the ends of their respective substrates (the tRNA or 5.8S rRNA) after cleavage by other enzymes. In eukaryotes the 5S rRNA is transcribed separately by RNA polymerase III. B. The Diplomonad Giardia lamblia has the usual order of rRNA subunits with short ITS regions, however RNase MRP has not yet been characterized from this species. RNAstructure folding of G. lamblia ITS1 [56] showing a single stranded region between two stem loops that could possibly be an A3 site. Other foldings of this sequence and foldings of other sequences (DQ157272 and AF239841) produce just a single stem-loop.;http://www.biomedcentral.com/
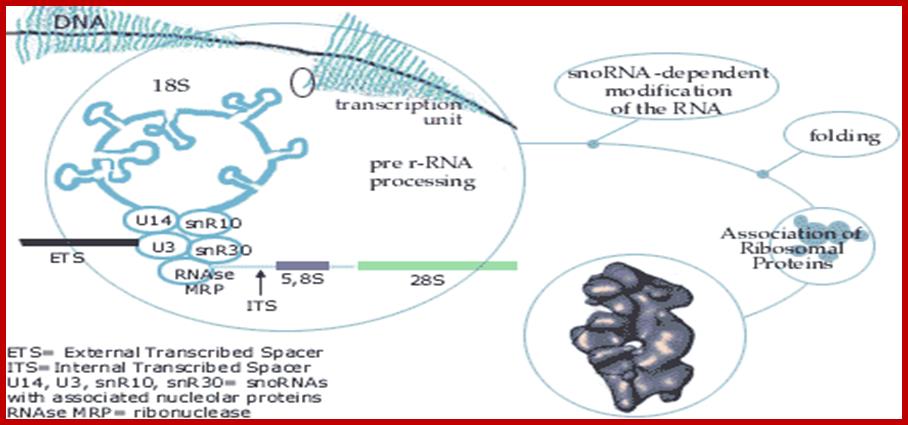
Prokaryotes contain ~ 70,000 ribosomes per cell. Eukaryotes contain more ribosomes approximately 8-10 million per cell, but human oocyte contains hundred times more than the normal number found in cells. Ribosomes in eukaryotes are larger 80S than prokaryotes 70S. Their genes are found in multiple copies and located only in certain chromosomes, far example in Eukaryotes their rRNA genes are found in secondary constriction region of chromosomes 13,14, 15 21 and 22. At the beginning of cell cycle chromosomal complex opens up and rDNA unwinds to allow transcription of rRNA gene which are organized in tandem repeats with intervals non transcribed space. The primary transcript is large and during initial folding snoRNA dependent cleavage of primary transcript takes place. Few other sno RNAs are involved modification of specific nucleotides in rRNA. This RNA now get associated with riboproteins in step by step manner, specific riboproteins by proteins assemble, while assembling 60s ribosome also get associated with another rRNA called 5S RNA. Process of transcription and assembly of proteins with processed rRNA takes place in a specific region called nucleolar organizer.
Assembly of ribosomal proteins with RNA and protein takes place in hierarchical mode.
http://nar.oxfordjournals.org/
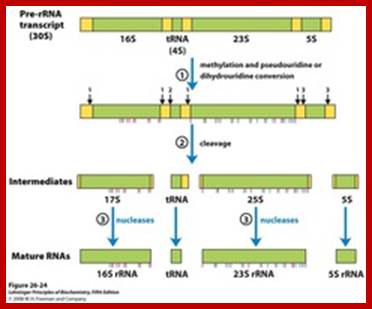
rRNA gene –precursor rRNA processing into smaller fraction; www.quzzlet,com; http://biosiva.50webs.org/
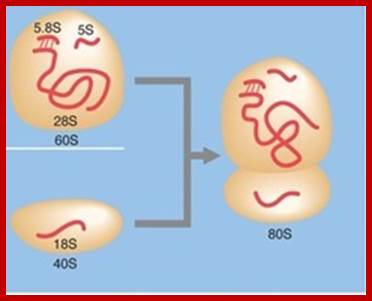
www.tumbler.com
Composition:
Compared to their prokaryotic homologs, many of the eukaryotic ribosomal proteins are enlarged by insertions or extensions to the conserved core. Furthermore, several additional proteins are found in the small and large subunits of eukaryotic ribosomes, which do not have prokaryotic homologs. The 40S subunit contains a 18S ribosomal RNA (abbreviated 18S rRNA), which is homologous to the prokaryotic 16S rRNA. The 60S subunit contains a 26S rRNA that is homologous to the prokaryotic 23S ribosomal RNA. In addition, it contains a 5.8S rRNA that corresponds to the 5' end of the 23S rRNA, and a short 5S rRNA. Both 18S and 26S have multiple insertions to the core rRNA fold of their prokaryotic counterparts, which are called expansion segments. For a detailed list of proteins, including archaeal and bacterial homologs please refer to the separate articles on the 40Sand 60S subunits.
|
Eukaryotic[3] |
Bacterial[3] |
||
|
Ribosome |
Sedimentation coefficient |
80 S |
70 S |
|
~3.2*106 Da |
~2.0*106 Da |
||
|
Diameter |
~250-300 Å |
~200 Å |
|
|
Large subunit |
Sedimentation coefficient |
60 S |
50 S |
|
Molecular mass |
~2.0*106 Da |
~1.3*106 Da |
|
|
Proteins |
47 |
33 |
|
|
rRNAs |
· 28 S rRNA (3354nucleotides) · 5 S rRNA (154 nucleotides) · 5.8 S rRNA (120 nucleotides) |
· 23S rRNA (2839 nucleotides) · 5S rRNA (122 nucleotides) |
|
|
Small subunit |
Sedimentation coefficient |
40 S |
30 S |
|
Molecular mass |
~1.2*106 Da |
~0.7*106 Da |
|
|
Proteins |
32 |
20 |
|
|
rRNAs |
· 18S rRNA (1753 nucleotides) |
· 16S rRNA (1504 nucleotides) |
https://en.wikipedia.org

www.biologyexams4u .com
Specific Ribosomal proteins and specific functions:
It is difficult to assign specific function to specific ribosomal proteins for all of them have cooperative action, yet some have specific functions. Ribosomes contain rRNA 28s, 5.8s and 18s rRNAs of about 120-4500 ntd long with 50-80 riboproteins of 25-300 a.a. in eukaryotes. Accuracy of transcriptions is in order of 1:20 000 to 1: 60 000 and translational accuracy is about is about 1:3000. E.coli ribosomes contain 5S rRNA (120ntds), 23s -2907 ntds and 16s-1542 ntds. Large ribosomes contain 36 proteins (L1-L36) and small subunit contains 21 subunits of proteins (S1-S21), these numbers derived form protein analysis on PAGE. All protein are single copy proteins with the exception of L7/L12. L7 is acetylated form of L12 and together with L10 organize into pentameric complex. Most of these protein’s tertiary structure has been established. Even the position of protein with respect to 3-D organizes rRNA. Sequential association is shown above in the page 12 and 13. Identified riboproteins , some are characteristic for all domains of life and some are characteristic proteins in bacteria, Archaea and eukaryotes. Ribosomal signatures, idiosyncrasies in the ribosomal RNA (rRNA) and/or proteins, are characteristic of the individual domains of life. One finds correlations between the rRNA signatures and signatures in the ribosomal proteins showing that the rRNA signatures coevolved with both domain-specific and universal ribosomal proteins (Ulrich Stelzl, M eta l;Korobeinikova. AV etal; Roberts etal).
S5 may also have a role in translocation
L2 and Peptide Bond Formation
Proteins of the Ribosomal Stalk: Factor Binding,
L11: binding of deacylated tRNA to the A site and the ‘GTPase associated centre’,
L9: Involved in the tRNA Arrangement at the P Site?,
Ulrich Stelzl; http://www.javeriana.edu.co/
Ribosome Mediated Inhibitors of translation:
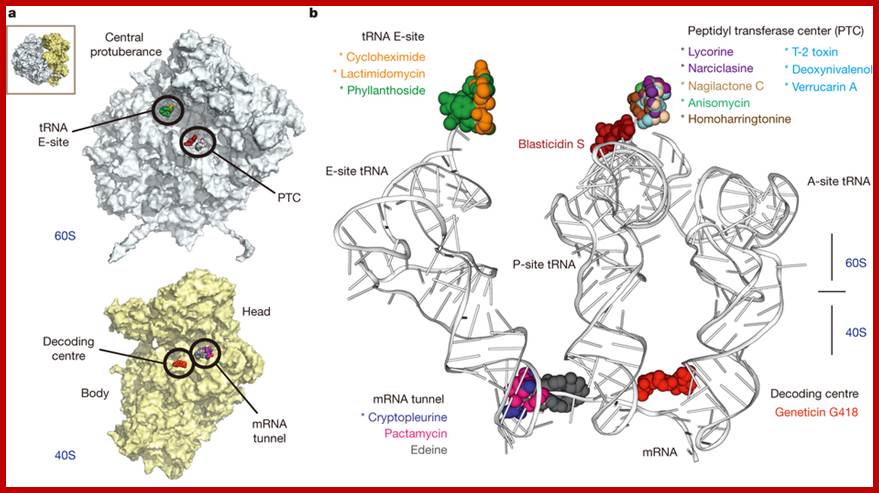
Binding sites of inhibitors on the yeast ribosome: The binding sites can be grouped in four functional regions: the tRNA E-site and the peptidyl transferase centre (PTC) on the large subunit (60S) and the decoding centre (DC) and the mRNA channel on the small subunit (40S). Twelve eukaryotic specific and 4 broadband spectrum inhibitors; all inhibitors were found associated with messenger RNA and transfer RNA binding sites. In combination with kinetic experiments, the structures suggest a model for the action of cycloheximide and lactimidomycin, which explains why lactimidomycin, the larger compound, specifically targets the first elongation cycle. The study defines common principles of targeting and resistance, provides insights into translation inhibitor mode of action and reveals the structural determinants responsible for species selectivity which could guide future drug development. Irina Prokhorova et al; http://www.nature.com/http://www.nature.com/
Kusugamycin: initiation (PK), displace F-met tRNA, mutants lack methylation of 16 s rRNA at the 3’end.
Streptomycin: initiation (PK), mutation in s12 of 30s ribosome causes resistance.
Kirromycin: elongation (PK), EF-Tu-GDP release is blocked by the antibiotic and no recycling.
Puromycin: elongation (PK), premature termination, because Puromycin has structure similar to tRNA configuration.
Erythromycin: peptidyl transfer (PK), blocks peptide bond formation, mutation in 23sRNA results in resistance.
Chloramphenicol: peptidyl transfer (PK), blocks peptidyl bond formation,
Cycloheximide: translocation (EK), inhibits peptidyl transferase on 60s subunit.
Fusidic acid: translocation (PK), EF-G-GDP cannot be released, no recycle.
Thiostrepton: translocation (PK) binds to 23sRNA and inhibits GTPase activity.