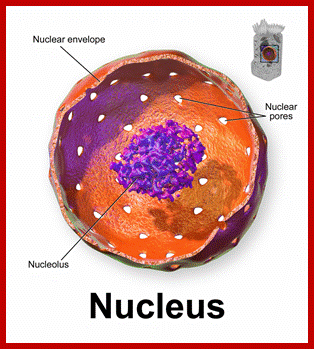
The Nucleus
1. Introduction
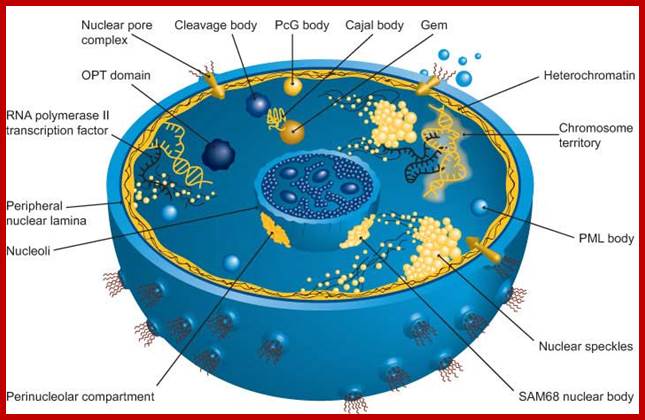
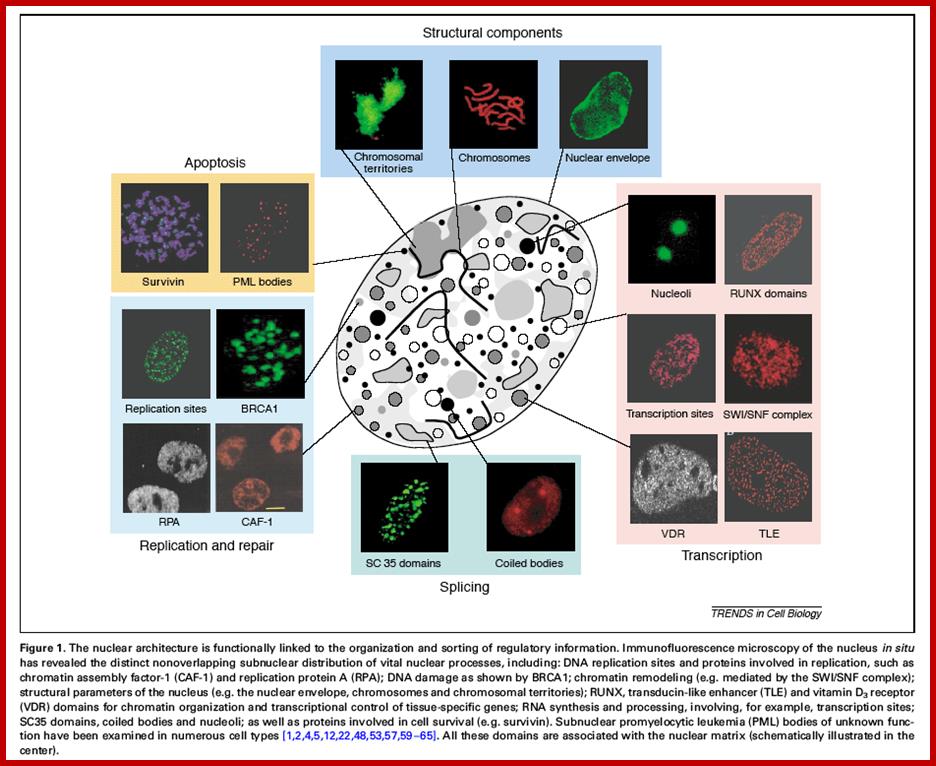
The nucleus is separated from the cytoplasm by two-unit double membranes. The outer nuclear membrane is continuous with the endoplasmic reticulum (Spector 2001; Lamond and Sleiman 2003). Exchange of proteins and mRNA between the cytoplasm and the nucleus occurs through multi-protein structures situated in the nuclear envelope known as nuclear pore complexes. The nucleus is compartmentalized and contains numerous sub-nuclear bodies, including nucleoli, Splicing speckles, Cajal bodies (CB), gems, and Promyelocytic leukemia (PML) bodies in addition to chromosomes. In contrast to cytoplasmic compartments, the sub-nuclear bodies lack a membrane separating them from the nucleoplasm. The buildup of factors in these distinct sub-nuclear bodies may serve to enhance the efficiency of specific nuclear functions.
 tp://www.dddarli.info/w
http://www.
tp://www.dddarli.info/w
http://www.
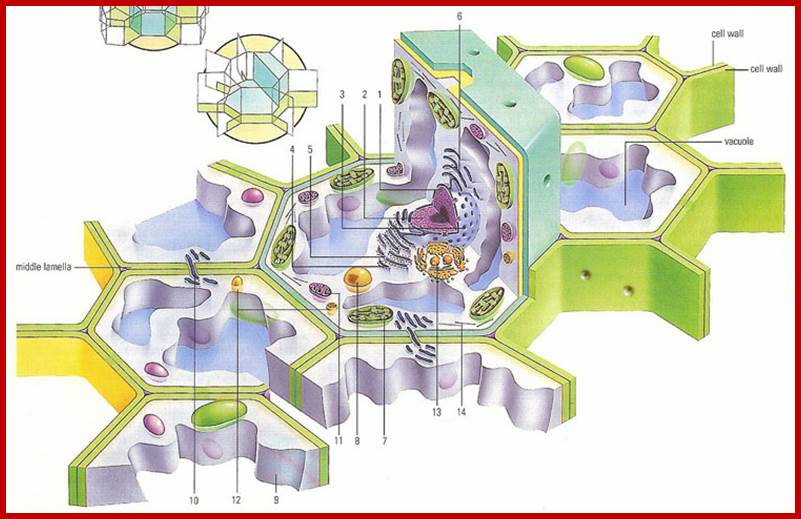
Cross section through a plant cell and surrounding cells; 1. Nucleus, 2. Chromatin, 3.; Nuclear membrane,4. ribosome, 5. Ribosomes anchored to ER. 6.Smooth ER, 7. Chloroplasts, 8. Enzyme containing microtubule, 9. Cytoplasm, 10. Plasmodesmata, 11. Mitochondria, 11. Lysosome, 12. 13. Golgi bodies and 14. Microfilaments and Microtubules; www.daviddarling.info
Nucleus:
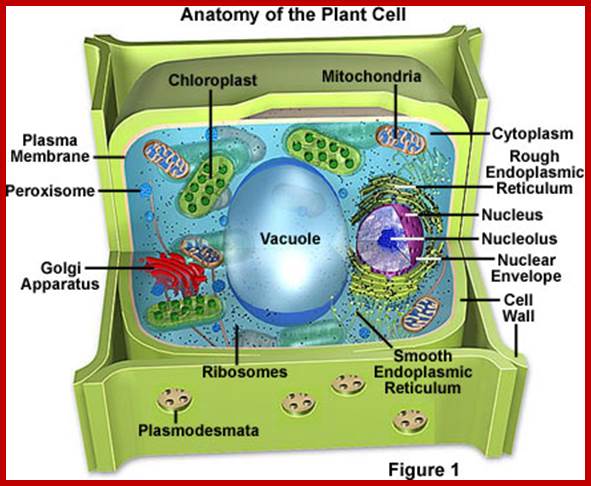
Nucleus is one of the most important organelles found in the cell, because it possesses all the genetic information necessary for inheritance, growth and development. Prokaryotes are lacking in well-defined nucleus, instead their genetic material is suspended freely in the protoplasm. On the contrary, in eukaryotic cells, the genetic material is highly organized into compact chromosomal structures. Furthermore, the chromosomes are localized within the nucleus and protected by a membrane.
https://en.wikipedia.org//Nucleolus
http://clinicalgate.com/nuclear-structure-and-dynamics/
https://micro.magnet.fsu.edu
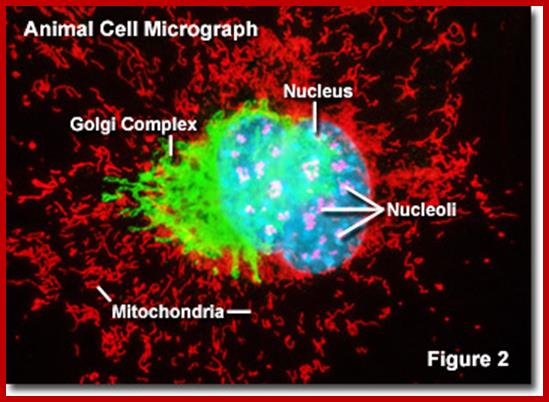
Fluorescent dye labeled Cell with its nucleus; https://en.wikipedia.org
Number and distribution:
Generally, the number of nuclei per cell is constant for a given species, but it may vary from species to species. Majority of plant and animal cells are monokaryotic, but in some plant species, the cells contain two nuclei per cell at a particular stage of development. Such a condition is referred to as dikaryotic condition; it may be homokaryotic or heterokaryotic. Examples - acrogenous hyphae in ascomycetes and secondary mycelium in basidiomycetes. However, in siphonales algae and phycomycetes fungi the cells are tubular and branched without any septa and contain a large number of nuclei. Such a condition is called coenocytic. The early liquid endosperm in coconut fruit is another example of multinucleate condition.
Nuclei in a coenocytic cell are normally distributed throughout the filamentous cells. But at the certain stages of development, they aggregate in a particular area for reproductive purpose. Ex: sporangia, gametangia. Otherwise, cells in most of the cases contain a single nucleus with no fixed position and it is always subject to movement by the underlying protoplasmic flux.
Shape: In most of the organisms, the shape of the nucleus is spherical; but it may assume slightly distorted shape transitorily due to the pressure exerted by cytoplasmic organelles. But in certain lower eukaryotes like ciliate protozoans the shape of nuclei vary from kidney shape to a string of beads. In such species, the shape is constant and characteristic of the given species.
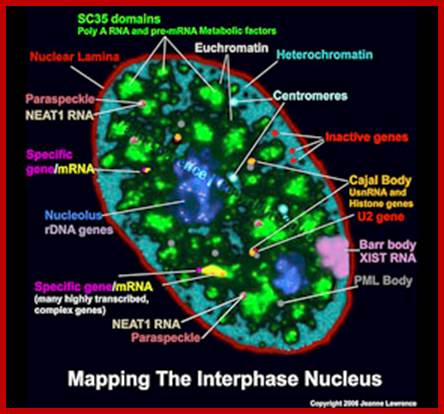
NEAT1 RNA is an architectural RNA that scaffolds a large and compartmentalized nuclear structure; NEAT1 RNA is a non-coding RNA that is required for the formation of paraspeckle (see image). Paraspeckle are ubiquitous nuclear structures (~10-30/nucleus) of unknown function found in all human primary and transformed cells.; http://www.umassmed.edu/
Bovine pulmonary artery endothelis cell labeled with probes to visualize nucleus, mitochondria and peroxisomes.;MitoTracker® Red CMXRos; https://www.thermofisher.com.
Cell mask with deep red plasma membrane stain: https://www.thermofisher.com
NEAT1 Nuclear Enriched Abundant transcriptr1 RNAs are not uncommon in Plants. Ex. Arabidopsis has this kind RNAs. Nuclear Enriched Abundant Transcript 1 (NEAT1) is a ~3.2 kb novel nuclear long non-coding RNA (RIKEN cDNA (2310043N10Rik). It is also known as Virus Inducible Noncoding RNA (VINC) or MEN epsilon RNA. It is transcribed from the multiple endocrine neoplasia locus. Expression of NEAT1 is induced in mouse brains during infection by Japanese encephalitis virus and rabies virus. NEAT1 is constitutively expressed in a number of non-neuronal tissues and cell lines. NEAT1 localizes to specific nuclear structures called Para speckles. NEAT1 RNA interacts with a Para speckle protein known as P54nrb or NONO and it is essential for Para speckle formation. Some studies demonstrate that NEAT1 RNA is essential for the formation and maintenance of Para speckles. Thus, this novel noncoding RNA appears to have an important structural role in the nuclear Para speckles.
http://www.sivabio.50webs.com/
Nucleolus:
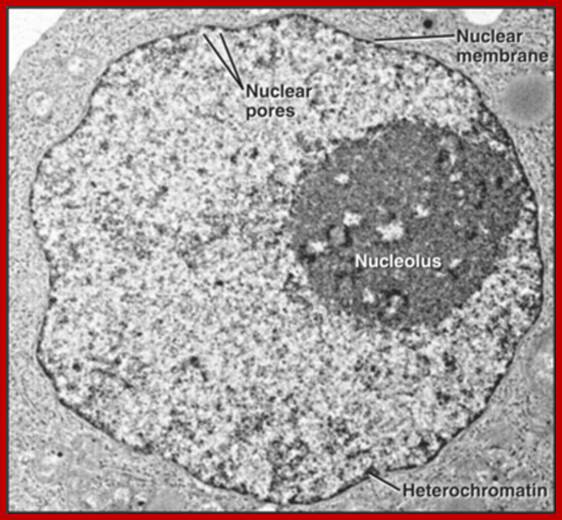
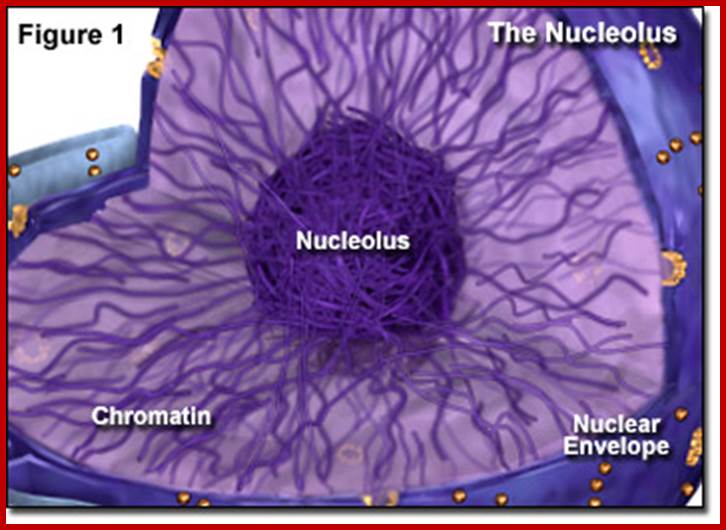
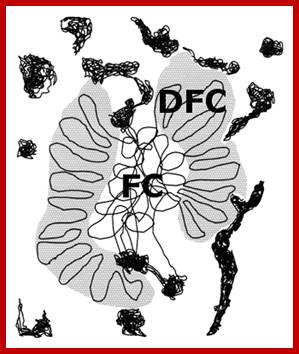
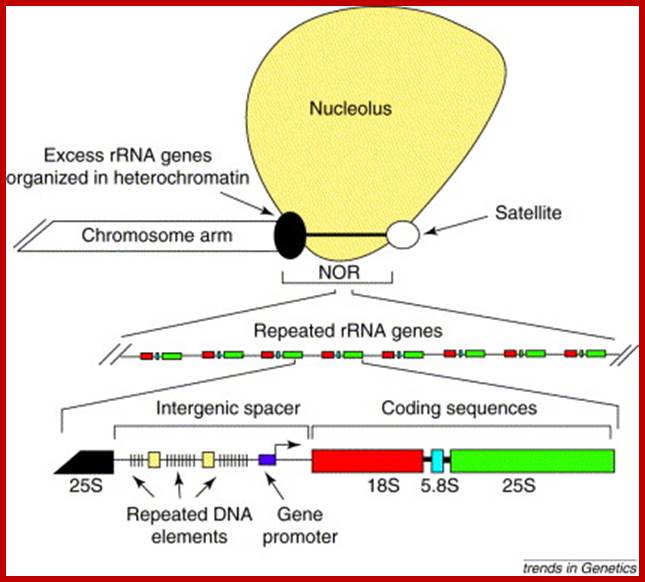
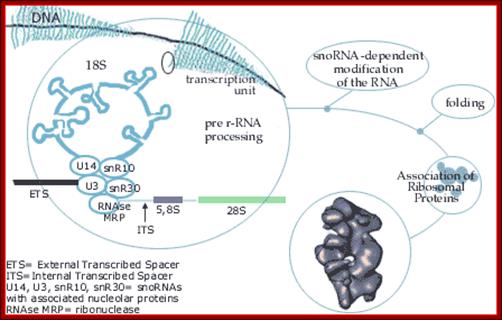
Most animal and plant cells contain 1-5 nucleoli, each 0.5-10 um (in wheat root cells) in diameter (Spector 2001; Lamond and Sleeman 2003; Zimber et al., 2004; Handwerger and Gall 2006). If the cell is active or activated the size of the nucleolus is 50-60 of the nuclear volume (Kantharaj et al). The nucleolus contains three distinguishable regions, the fibrillar center (FCs)- pars fibrosa, which is surrounded by the dense fibrillar component (DFC) and the granular component (GC) pars granulosa that constitutes the rest of the nucleolus-pars amorpha. Nucleoli form around tandemly repeated clusters of ribosomal RNA (rRNA) genes which opened up rRNA DNA and their transcripts. These loci are termed nucleolar organizer regions (NORs) first described by Barabara McClintok. The function of the nucleolus is to process precursor RNAs (45s) copied by RNA pol I and RNAP III and import ribo-proteins and assemble ribosomal subunits. The rRNA genes are transcribed by RNA polymerase I as a large pre-rRNA precursor with ITS and ETS that is cleaved to produce 5.8S, 18S, and 28S rRNAs found in ribosomes and the 5s RNA is found in the same region or at different sites in chromosomes. rRNA is post-transcriptionally modified by guide RNAs (belong to a class of sno RNAs) at specific sites and assembled with ribosomal proteins, which are synthesized in the cytoplasm and imported into the nucleus through the nuclear pores. This results in the formation of the large and small ribosomal sub-units, which are subsequently transported from the nucleus to the cytoplasm where they mediate mRNA translation. Nucleolus also contains a central clear region called nucleolar vacuole. Once ribosomes are assembled they are transported across Nuclear pore complex into cytoplasm.
http://micro.magnet.fsu.edu/
The ultra-structure of
nuclei reveals that they contain many small bodies organized to perform various
functions. Among them the nucleolus is the largest. Among others cajal bodies
and nuclear speckles are important.
Most animal
and plant cells contain 1-5 nucleoli, each 0.5-5 um in diameter (Spector 2001; Lamond
and Sleeman 2003; Zimber et al., 2004; Handwerger and Gall 2006). The nucleolus contains three distinguishable
regions, the fibrillar centers (FCs), which are surrounded by the dense
fibrillar component (DFC) and the granular component (GC) that constitutes the
rest of the nucleolus. Nucleoli form around
tandemly repeated clusters of ribosomal RNA (rRNA) genes. These loci are termed
nucleolar organizer regions (NORs). The function of the nucleolus is to synthesize rRNA and assemble
ribosomal subunits. The rRNA genes are transcribed by RNA polymerase I as a
large pre-rRNA precursor that is cleaved to produce 5.8S, 18S, and 28S rRNAs
found in ribosomes. rRNA is post-transcriptionally modified and assembled with
ribosomal proteins, which are synthesized in the cytoplasm and imported into
the nucleus through the nuclear pores. This
results in the formation of the large and small ribosomal sub-units, which are
subsequently transported from the nucleus to
the cytoplasm where they mediate mRNA translation.
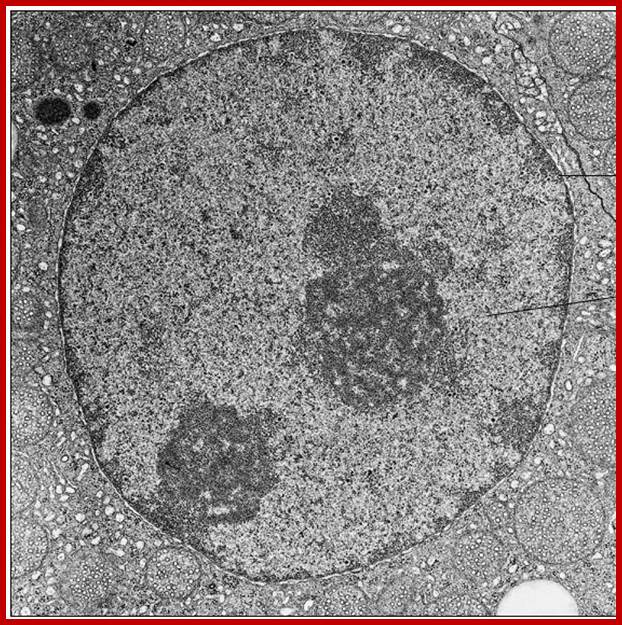
EM of Perikaryon of a nerve cell in a spinal Ganglion:www.netterimages.com
Nuclear Satellite-Nucleolar Mangalyanum
The polar thickness map, as superposed on the nucleus. Color represents thin (dark-red) peripheric chromatin, mostly associated with the nuclear envelope. Thick, bulky zones penetrate into the core of the nucleolus (light-yellow) (see Plate10). The apparent higher resolution (compare with the surface renderings) comes from the 3D-Bresenham traversal of the volume, which is done at a sub-voxel resolution, and averaging measurements for depth values as well as surface intersections. http://www.academicos.ccadet.unam.mx/
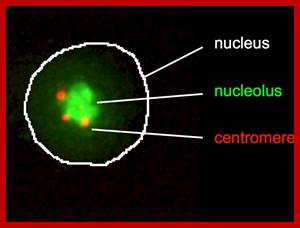
Centromeres cluster at the periphery of the nucleolus; The nucleolus protein modulo anchors the complex consisting of the centromere and NLP to the nucleolus, and the protein CTCF supports NLP in the clustering of the centromeres. Patrick Heun et al https://www.ie-freiburg.mpg.de
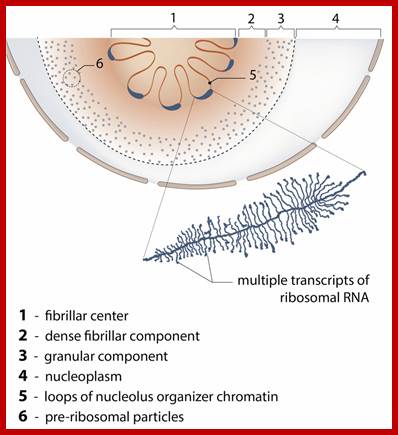
One segment of rRNA gene and gene transcript; http://www.mechanobio.info/
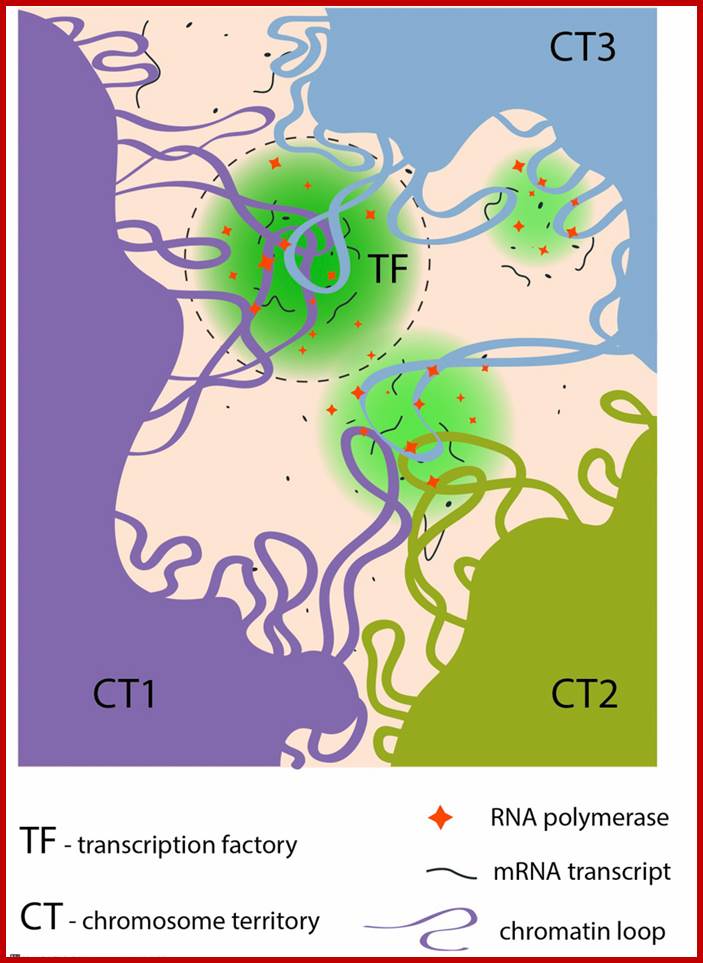
Certain regions of chromatin are bundled together where transcription goes on and the region appears as if a factory.; Transcription Factories. ; http://www.mechanobio.info/
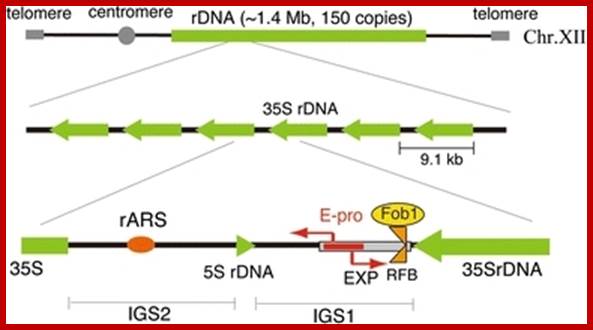
Structure of the budding yeast rDNA locus. The rDNA is a tandem repeating array on chromosome XII. A repeating unit (9.1 kb) has 5S and 35S rRNA genes and two intergenic spacer regions (IGS1, 2). rARS and RFB are the replication origin and replication fork barrier site, respectively. EXP (~500 bp) is an expansion sequence that contains RFB and E-pro. E-pro is a bidirectional promoter for non-coding transcripts that function in the regulation of rDNA repeat numbers. The rDNA structure is broadly conserved from yeast to human, though in the human genome the 5S rDNA is found in independent arrays; In budding yeast one can observe the location of 5srRNA; ; https://openi.nlm.nih.gov.
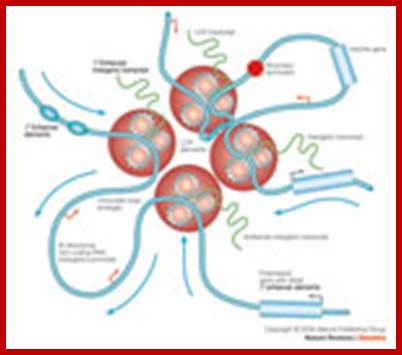
Replication and transcription: Shaping the landscape of the genome;;Lyubomira Chakalova et al;www.nature.com; Revealing the unseen; Nucleoli the hub for unwinding chromatin DNA loops for rRNA transcription; Marco Biggiogera et al; jcs.biologists.org
Nucleus showing nuclear membrane with nuclear pore complexes, nucleolus and inner nuclear lamellae proteins associated with chromatin; www.rumalis.com
The nucleolus as sub nuclear compartment was first observed more than 200 years ago in which takes place transcription of ribosomal RNA (rRNA) genes and the assembly of ribosomes. As an average mammalian cell can produce up to 10,000 ribosomes per minute, cells have to invest a very large portion of their own metabolic effort to meet demand from protein synthesis Dr. Raffaella Santoro.
Nucleolus is a sub nuclear structure found as rRNA rich with its proteins; this is where ribosomes are generated. The number of nucleoli is one, where smaller nucleolar fragments fuse to produce a large structure. All nucleoli generate from secondary constriction of chromosomes-called nucleolar organizers. This is where the rRNA genes located, when nuclear membrane reforms at the end of telophase the secondary constriction region becomes active and rRNA genes unfold and extend in to the nuclear sap and the rRNAs are transcribed from multisubunit-repeats of rRNA genes (can be 50to 100) by RNA polymerase-I. This region can be observed as dense mass of RNA and protein complex.

Repeats of rRNA genes with spacers; www.edoc.hu-berlin.de
Epigenetic phenomena Epigenetic process is/are heritable, alternative states of gene activity that are not explained by mutation, changes in gene sequence or normal developmental regulation. Among the earliest examples was nucleolar dominance, a common phenomenon in interspecific hybrids in which only ribosomal RNA (rRNA) genes inherited from one parent are transcribed. Only active rRNA genes initiate formation of a nucleolus, hence the name for the phenomenon. As in other epigenetic phenomena, chromatin modifications enforce selective gene silencing in nucleolar dominance. However, the mechanisms that discriminate between parental sets of rRNA genes are unclear. Possibilities include sequence differences that affect transcription factor affinities. Other evidence suggests that chromosomal context is more important than rRNA gene sequences, implying control on a larger scale (Scince.com).
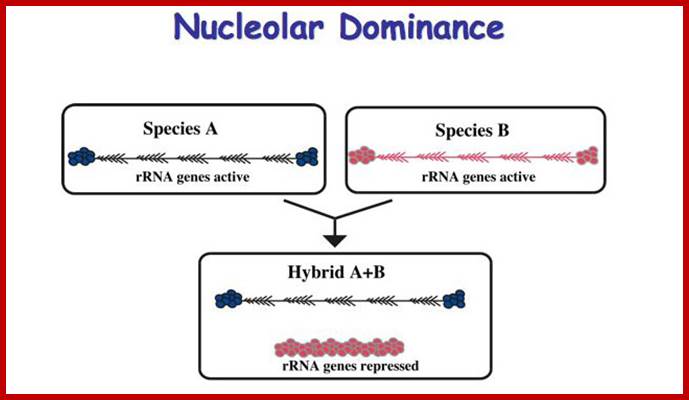
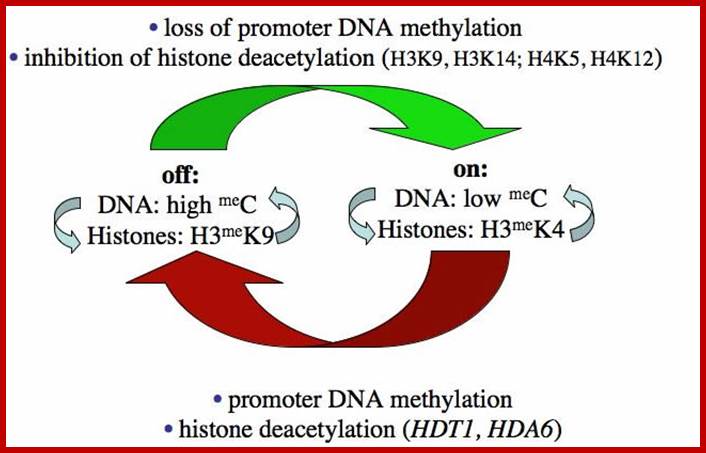
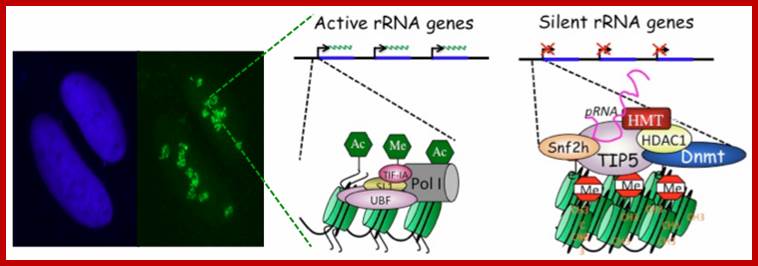
Such type of behavior is due to selective silencing of one set by Histone methylation or/and Histone deacetylation or both. This is called epigenetic switch on or off that uses RNAi for rRNA processing.
Epigenetics of nucleolar dominance ;http://www.sciencedirect.com/
NEAT1 Nuclear Enriched Abundant transcriptr1 RNAs are not uncommon in Plants. Ex. Arabidopsis has this kind RNAs. Nuclear Enriched Abundant Transcript 1 (NEAT1) is a ~3.2 kb novel nuclear long non-coding RNA (RIKEN cDNA (2310043N10Rik). It is also known as Virus Inducible Noncoding RNA (VINC) or MEN epsilon RNA. It is transcribed from the multiple endocrine neoplasia locus. Expression of NEAT1 is induced in mouse brains during infection by Japanese encephalitis virus and rabies virus. NEAT1 is constitutively expressed in a number of non-neuronal tissues and cell lines. NEAT1 localizes to specific nuclear structures called paraspeckles. NEAT1 RNA interacts with a paraspeckle protein known as P54nrb or NONO and it is essential for paraspeckle formation. Some studies demonstrate that NEAT1 RNA is essential for the formation and maintenance of paraspeckles. Thus, this novel noncoding RNA appears to have an important structural role in the nuclear paraspeckles
The ultra-structure of
nuclei reveals that they contain many small bodies organized to perform various
functions. Among them the nucleolus is the largest. Among others cajal bodies
and nuclear speckles are important.
Most animal
and plant cells contain 1-5 nucleoli, each 0.5-5 um in diameter (Spector 2001;
Lamond and Sleeman 2003; Zimber et al., 2004; Handwerger and Gall 2006). The nucleolus contains three distinguishable
regions, the fibrillar centers (FCs), which are surrounded by the dense
fibrillar component (DFC) and the granular component (GC) that constitutes the
rest of the nucleolus. Nucleoli form around
tandemly repeated clusters of ribosomal RNA (rRNA) genes. These loci are termed
nucleolar organizer regions (NORs). The function of the nucleolus is to synthesize rRNA and assemble
ribosomal subunits. The rRNA genes are transcribed by RNA polymerase I as a
large pre-rRNA precursor that is cleaved to produce 5.8S, 18S, and 28S rRNAs
found in ribosomes. rRNA is post-transcriptionally modified and assembled with
ribosomal proteins, which are synthesized in the cytoplasm and imported into the nucleus through the nuclear pores. This
results in the formation of the large and small ribosomal sub-units, which are
subsequently transported from the nucleus to
the cytoplasm where they mediate mRNA translation.
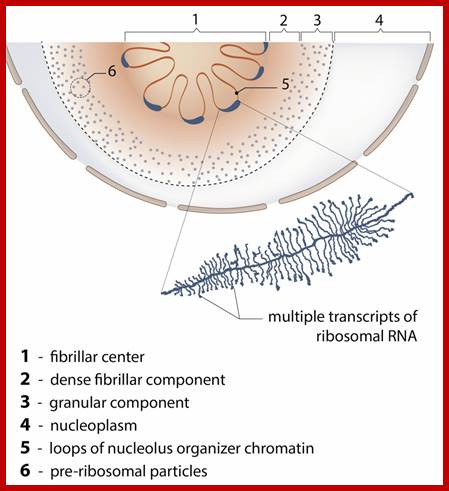
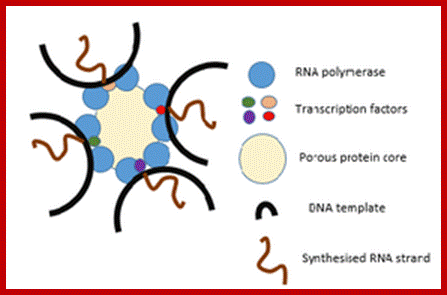
In this site or locus more number of genes are transcribed and they are clubbed together; https://en.wikipedia.org/

Schematic diagram of transcription of multiple genes at a nuclear RNAPII transcription factory. RNAPII factory shown as central blue circle with three transcribing genes and their associated transcription factors (small colored circles). Nascent transcripts are shown in red, chromatin is dark blue, and splicing components are depicted as small black circles with orange halo. http://cshperspectives.cshlp.org/
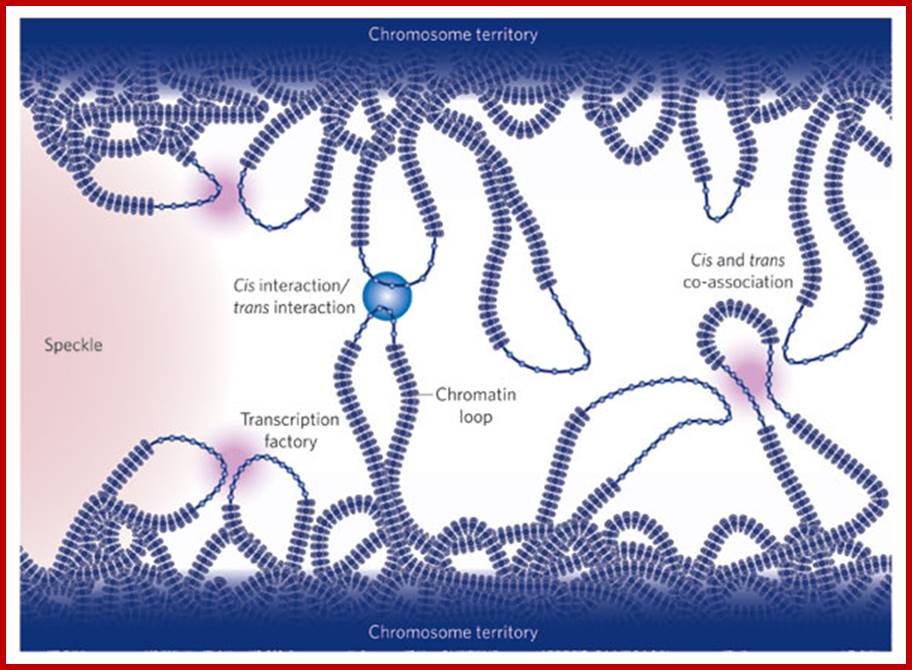
Active genes on decondensed chromatin loops that extend outside chromosome territories can colocalize both in cis and in trans at sites in the nucleus with local concentrations of Pol II (namely transcription factories; dark pink) and adjacent to splicing-factor-enriched speckles (pale pink). Interactions can also occur between regulatory elements and/or gene loci and lead to coregulation in trans (blue circle). http://www.nature.com/
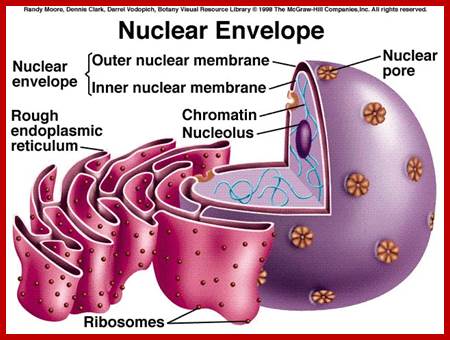
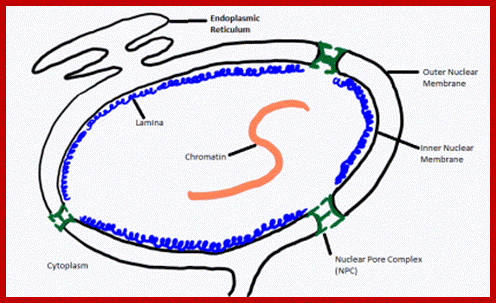
Nuclear Membrane:
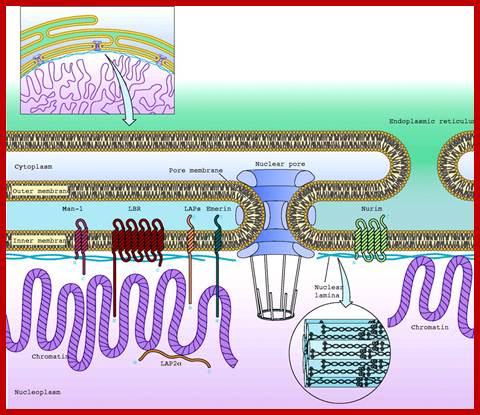
The presence of nuclear envelope around the genetic material distinguishes the eukaryotes from that of prokaryotes. The nuclear envelope consists of an outer membrane and an inner membrane separated by a space called perinuclear space.
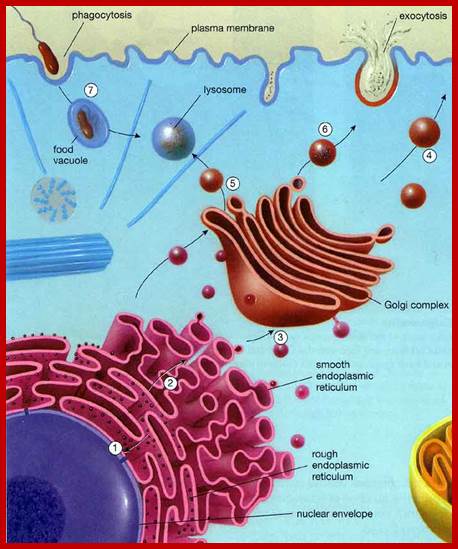
Nuclear outer membrane is in contact with ER. Which is in relation with endo and exo vesicles;.www.uic.edu
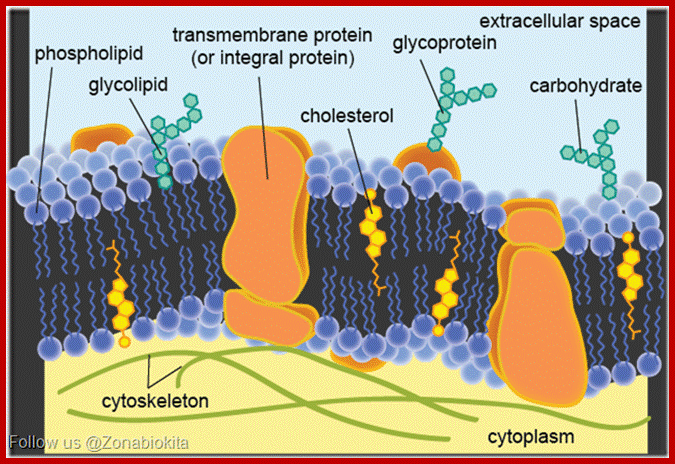
Nuclear membrane.http: //www.zonabiokita.blogspot.in/
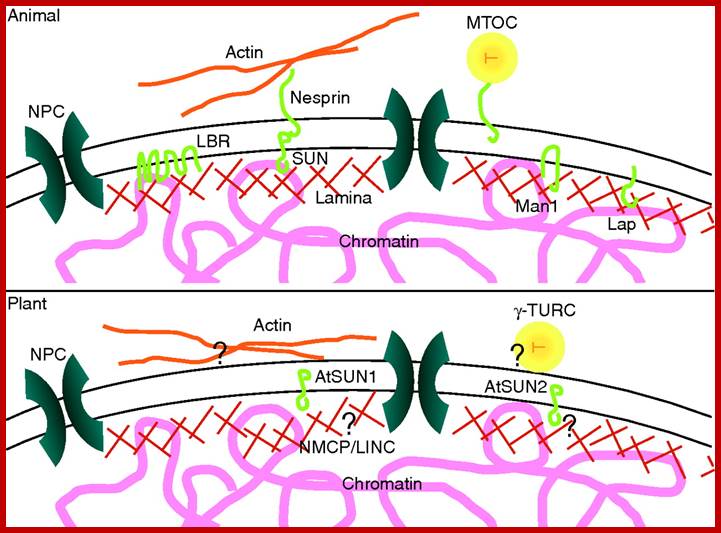
The nuclear envelope in the plant cell cycle: structure, function and regulation; Scaffold-structures support the nuclear envelope. In both animals and plants, the nuclear envelope is associated with nucleoskeletal and cytoskeletal structures. In animals the lamina is connected to the inner nuclear membrane by membrane-intrinsic proteins such as LBR, Man1, SUN domain proteins and Laps, which also associate with chromatin. KASH-domain proteins such as nesprins bind to SUN domain proteins and stretch into the cytoplasm to link to cytoskeletal elements such as actin and microtubule organizing centers (MTOC). In plants, a lamina-like network is also present and is hypothesized to consist of filamentous proteins such as NMCP1/2 and LINC1/2. How these proteins and the meshwork are associated with the nuclear envelope remains to be established but AtSUN1 and AtSUN2 are putative anchor candidates. Cytoskeletal structures such as actin and gamma tubulin ring complexes (γ-TURC) are associated with the cytoplasmic face of the plant nuclear envelope but similarly their anchoring mechanisms remain unknown.; www,aob.oxfordjournals.org
At the outer surfaces of the outer membrane, many ribosomes are found attached and they are actively engaged in protein synthesis. It is possible that some of the proteins synthesized in such sites may find their way into the nucleus by facilitated transport. The inner surface of the inner membrane is associated with intermediate fibers called lamins, they form a kind of reticular network and provide mechanical support to the membrane. Chromosomes are found bound to the inner surface of the membrane.
It is filled with a fluid containing same granular structures and enzymes. The outer nuclear membrane is in continuity with endoplasmic reticulum. In fact, the outer membrane continuously produces numerous finger shaped outgrowths, which expand into flat membranous sacs. This process is called blebbing, which is very common in some protozoan nuclei
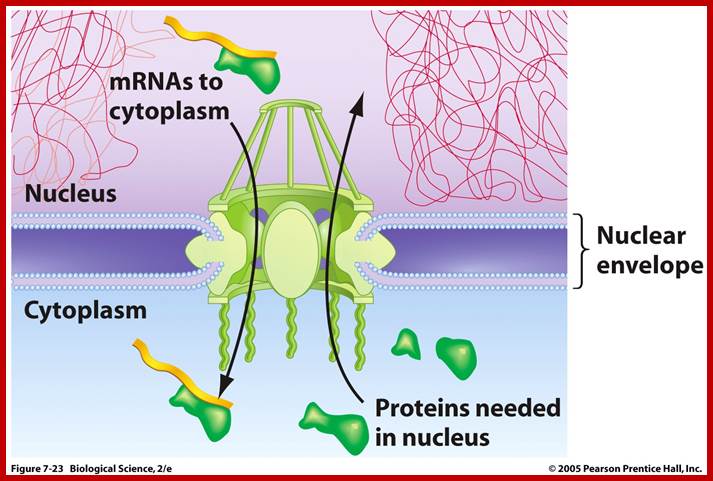
Nuclear pore complex:
Nuclear membrane is not a continuous sheet but is interrupted by a number of minute openings called nuclear pores through which materials pass through from the nucleus to cytoplasm and vice versa. The number of pores present per cm2 varies from species to species and it further depends upon the physiological state of the cell.
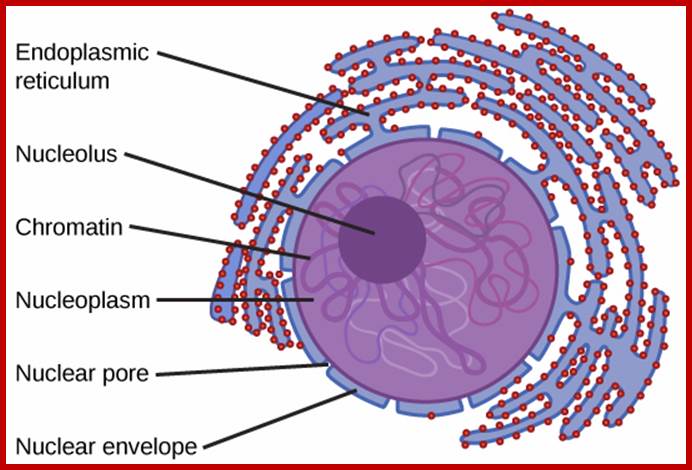
The outermost boundary of the nucleus is the nuclear envelope. Notice that the nuclear envelope consists of two phospholipid bilayers (membranes)—an outer membrane and an inner membrane—in contrast to the plasma membrane (Figure), which consists of only one phospholipid bilayer. (credit: modification of work by NIGMS, NIH); http://cnx.org/
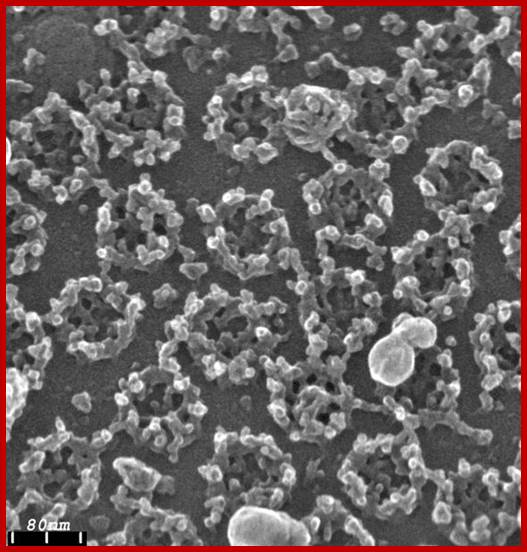
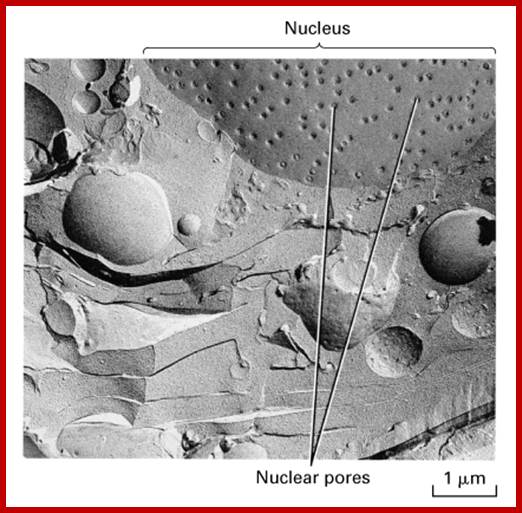
http://www.leica-microsystems.com/ Field emission scanning electron micrograph of an isolated Xenopus laevis oocyte nuclear envelope showing the outer surface the outer nuclear membrane and the nuclear pore complexes.
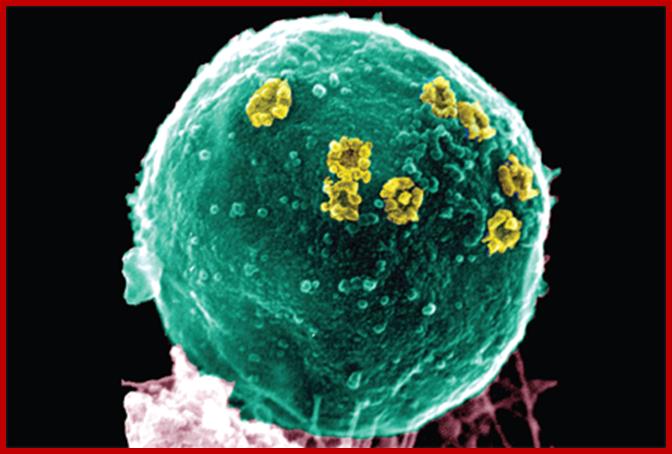
Scanning electron micrograph of nuclear pore complexes (yellow) in the nuclear envelope of S. cerevisiae. Image is from the protocol by Kiselevaet al. Cover by Jessica Iannuzzi.http://www.nature.com/
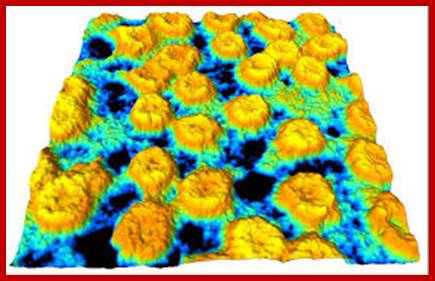
Nuclear pore complexes; www.stolaf.edu
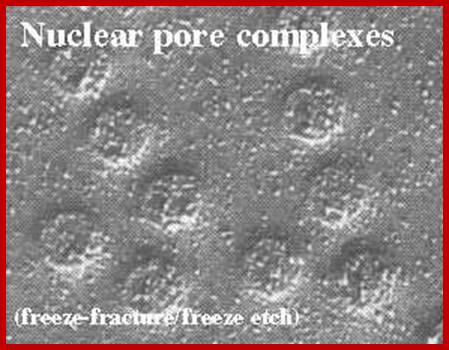
Freeze fractured etch Photo-http://cytochemistry.net/cell-biology
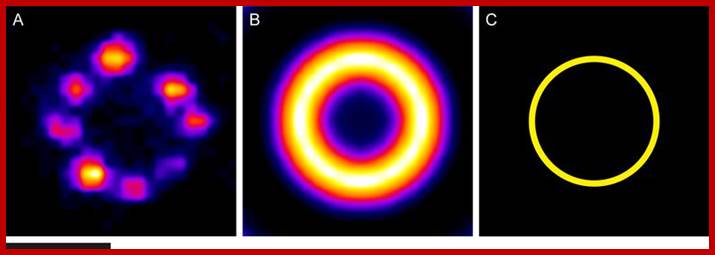
Example results of the averaging procedure. (A) An image of an NPC labeled with an antibody against Nup96 (Gaussian filtered). (B) An average image of NPC stained with an antibody against Nup96 generated by alignment and summing of over 8,000 images of individual pores. (C) The average position of fluorescent markers on Nup96 around the center of the NPC, determined from the profile of the average image. Scale bar: 0.1 μm. http://www.leica-microsystems.com/science-lab

http://www.leica-microsystems.com/science-lab
ig. 3: Applying the averaging procedure to several epitopes of the Nup107–160 subcomplex (depicted by different colors), their relative positions can be determined. Staining was achieved by using nanobody labeling of GFP fusion proteins. Positional information was overlaid with the EM structure of the cytoplasmic ring of the NPC. Electron density map courtesy of Prof. O. Medalia ].
Analysis of nuclear architecture and chromatin structure following synaptic activation in vivo and in vitro;
A second research theme in my laboratory is centered on the identification of neuronal genes that are regulated in vivo following exposure to Novel Enriched Environmental (NEE) conditions, a complex somatosensory mode of stimulation.. Prolonged exposure to NEE protects against neuro-degeneration; it also enhances neurogenesis, dendritic arborization and resistance to apoptosis.
http://www.ucl.ac.uk/
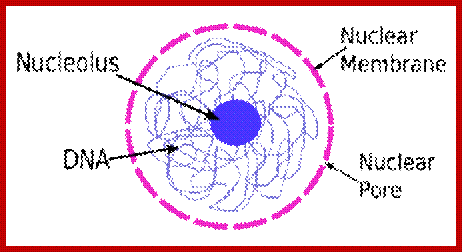
Nucleus; http://study.com
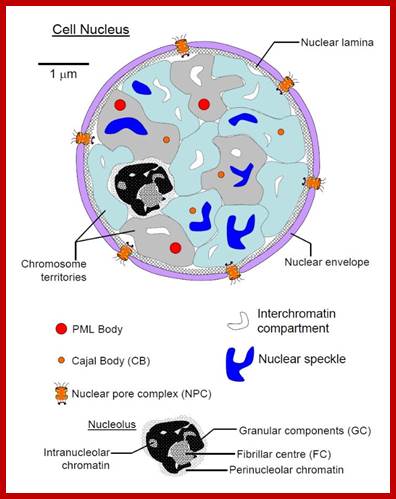
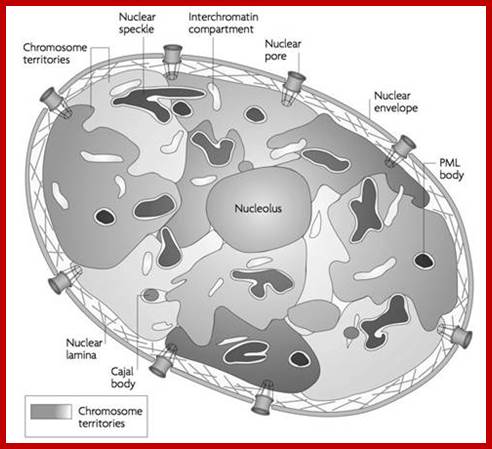
The diagram above illustrates some additional structures found inside the nucleus. Inside the nuclear envelope is a network of protein fibres called the nuclear lamina, made of the protein laminin. The nuclear lamina is part of the cytoskeleton and supports the nuclear membrane. Cajal bodies are 0.1-2 micrometres in diameter and number 1-10 per nucleus and they are involved in the manufacture of certain specialised RNA molecules. The function of the PML bodies is unknown. Each chromosome territory corresponds to the region occupied by the chromatin of a single chromosome. In a human body cell there are 46 chromosomes (23 pairs, 23 paternal and 23 maternal); http://cronodon.com/BioTech/Cell_Nucleus.html
Initially genes regulated were identified following NEE conditions, by using ChIP Seq assay, a technique that combines chromatin immunoprecipitation with large-scale direct ultrahigh-throughput DNA sequencing. Analysis of H3K9/K14 acetylation (a chromatin mark enriched in promoter regions of actively transcribed genes) of somatosensory cortex coupled with microarray analysis revealed that following exposure to NEE conditions neurons undergo a reactivation of a developmental transcriptional program. Moreover, an hyper-acetylated and highly conserved region was significantly enriched within the promoters of genes induced following NEE stimulation. We have now evidence that this sequence mediates chromatin tethering and recruitment to transcription factories of genes that are co-transcribed following synaptic stimulation.
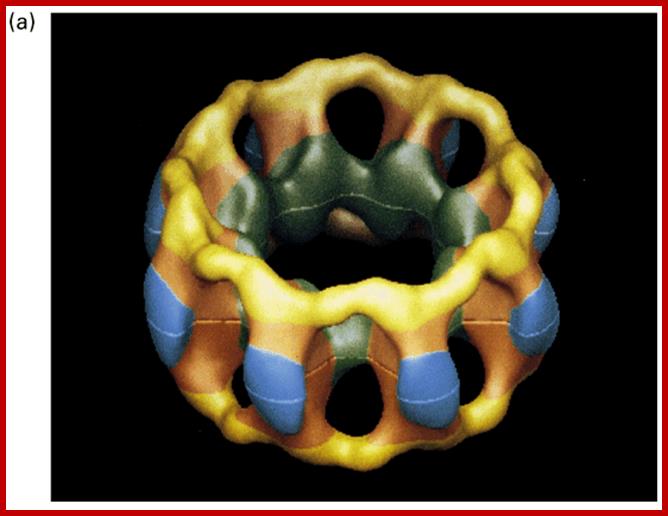
Nuclear pore complex; http://www.pha.jhu.edu/
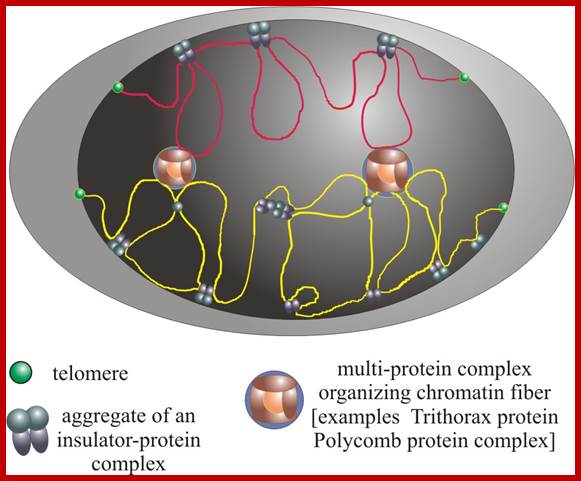
Nucleus and chromatin territorial structure jpg; https://upload.wikimedia.org
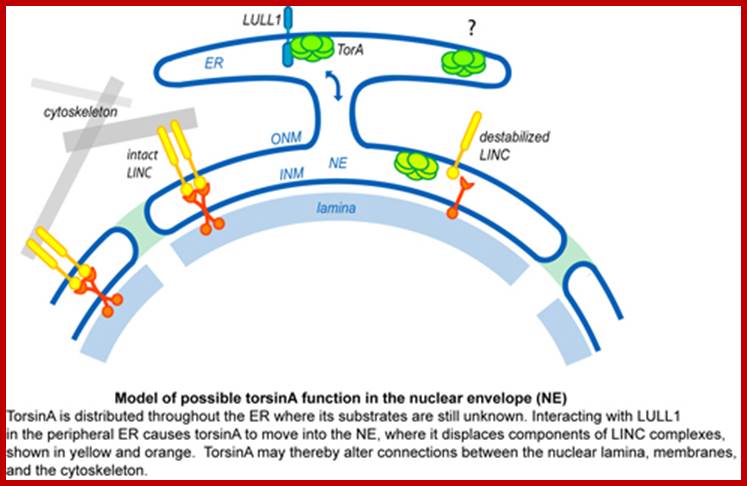
ttp://www.hansonlab.wustl.edu/
TorsinA (TorA) is an AAA+ ATPase in the endoplasmic reticulum (ER) lumen that is mutated in early onset DYT1 dystonia a neurological movement disorder (limbs). TorA-an AAA+ATPase ( LULL1) is also known as TOR1AIP) is an essential protein in mice and is thought to function in the nuclear envelope (NE) despite localizing throughout the ER. Here, we report that transient interaction of TorA with the ER membrane protein LULL1 targets TorA to the NE. FRAP and Blue Native PAGE indicate that TorA is a stable, slowly diffusing oligomer in either the absence or presence of LULL1. Increasing LULL1 expression redistributes both wild-type and disease-mutant TorA to the NE, while decreasing LULL1 with shRNAs eliminates intrinsic enrichment of disease-mutant TorA in the NE. When concentrated in the NE, TorA displaces the nuclear membrane proteins Sun2, nesprin-2G, and nesprin-3 while leaving nuclear pores and Sun1 unchanged. Wild-type TorA also induces changes in NE membrane structure. Because SUN proteins interact with nesprins to connect nucleus and cytoskeleton, these effects suggest a new role for TorA in modulating complexes that traverse the NE. Importantly, once concentrated in the NE, disease-mutant TorA displaces Sun2 with reduced efficiency and does not change NE membrane structure. Together, our data suggest that LULL1 regulates the distribution and activity of TorA within the ER and NE lumen and reveal functional defects in the mutant protein responsible for DYT1 dystonia.; INM-P Inner nuclear membrane proteins, ttp://www.hansonlab.wustl.edu/
Acronyms=LULL1, retard TorsionA to nuclear envelope revealing that is impaired by DYT1 dystonin mutation; LULL1 stands for luminal domain like LAP1 protein; FRAP-fluorescence recovery after bleaching used in studying molecular dynamics in living cells. SUN- Sad1-UNC-84 homology) domain-molecular bridge in nuclear envelope; The LINC (linker of nucleoskeleton and cytoskeleton) complex, formed by the SUN and the nesprin proteins at the nuclear envelope, The SUN domain is sufficient to mediate binding to the KASH (Klarsicht, ANC-1, and Syne homology) domain of nesprin 2.
Laminar elements at the inner surface of nuclear membrane; Lamins; https://cellbiology.med.unsw.edu.au
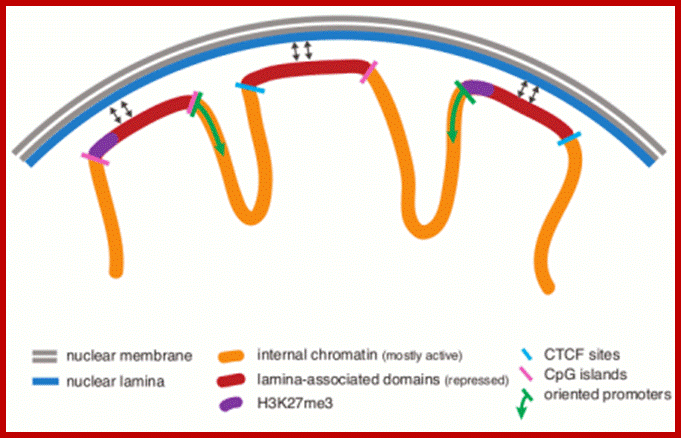
Many chromosomal domains are associated with nuclear lamibna, demarcated by putative insulators; http://epigenome.cbrc.jp/
http://www.cbi.pku.edu.cn/
A representation of the nucleus showing the location of lamins and actin and some of their proposed interactions. Lamins are represented by thick red lines near the nuclear envelope, indicating a concentration in the lamina, and as thin red lines throughout the nucleoplasm. Some inner nuclear membrane lamin-binding proteins are shown (LAP1, LAP2b, emerin and Syne/Anc-1 ). An example of a lamin interaction with nuclear pores is shown for NUP153. Proposed functional roles for lamins in large complexes are indicated in the two chromosome domains (indicated in blue and light brown). Lamins are associated with both the DNA replication complex containing DNA polymerase d, RFC and PCNA; the transcription complex containing pol II and TFIID; and spliceosomes. The interaction of actin with the spliceosome through heterogeneous ribonucleoproteins (hnRNPs) and small nuclear ribonucleoproteins (snRNPs) is also shown. The proposed functions of actin and Arps in chromatin remodeling is indicted by its interaction with the BAF complex (large yellow area). LAP2 and emerin share a 40-residue homology domain called LEM (pink box) that binds to BAF, the barrier to autointegration factor. Through BAF, lamins can interact with chromatin. Protein 4.1 is shown binding to dimerized Syne/Anc-1 and actin. Syne/Anc-1 is also shown to form a complex with emerin, indicating another potential link between the lamina and actin. ( ) Syne/Anc-1 has many names in the published literature, including Myne, Nesprin and NUANCE, and is related to a Drosophila protein MSP300. (See text for full details.) This figure is adapted from [1], and reproduced with permission from Cold Spring Harbor Laboratory Press, copyright 2002.
![]()

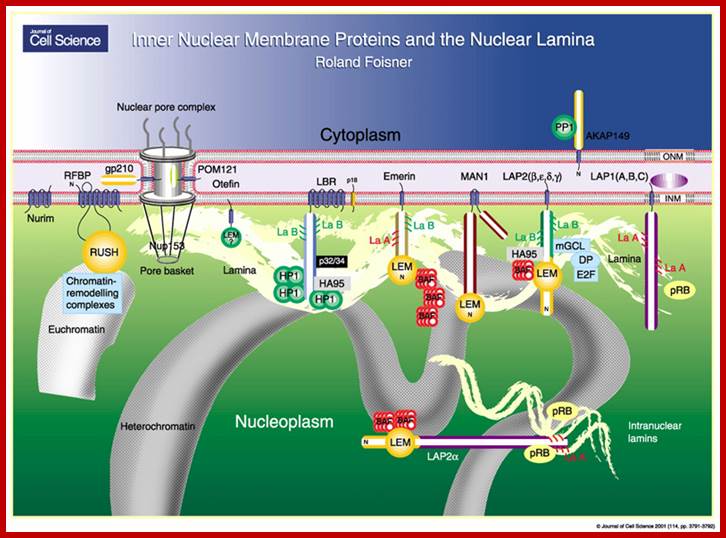
The lamins bind to specific proteins present in the inner nuclear envelope, such as emerin and lamin B receptors. The lamins provide skeletal support to nuclear envelope. These also provide the site for attachment to chromatin fibers. Lamins also help in dissolution of nuclear envelope at the time of cell division and its reorganization after the cell division is over. When phosphate groups are not attached to lamins, lamins are assembled to form an ordered structure, which help in organizing nuclear envelope. Moreover, at the time of cell division, lamins are phosphorylated resulting in their depolymerization so that nuclear envelope also disappears. The phosphorylation and dephosphorylation of lamins is triggered by a specific protein kinase.

Nuclear Lamina, a fibrous network of proteins; at inner surface of the Nuclear membrane consists of different mol.wt called A,B1 and C- mol.wt is 60-80kDa, they are fibrous related intermediate proteins; formation of theis structure at the inner surface provides support and strength.; http://kc.njnu.edu.cn/
Proteins called Lamins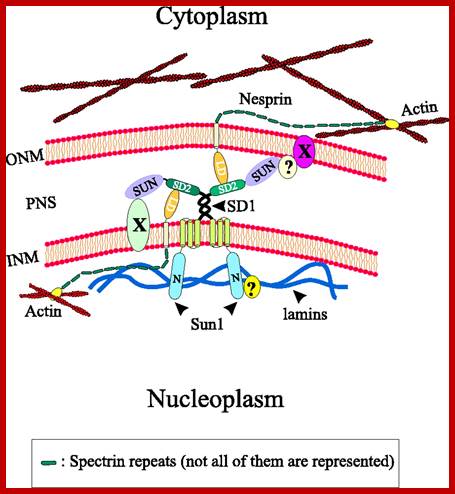
Model illustrating the interactions of Sun1 with Nesprins at the nuclear envelope. Unknown nuclear envelope proteins and interactions are indicated by X and?, respectively. To reduce complexity a homotypic dimerisation of Sun1 via the coiled-coil regions is postulated, although other coiled-coil-containing proteins might form heterotypic complexes with Sun1. INM, inner nuclear membrane; LD, luminal domain; N, N-terminal domain; ONM, outer nuclear membrane; PNS, perinuclear space. http://jcs.biologists.org/
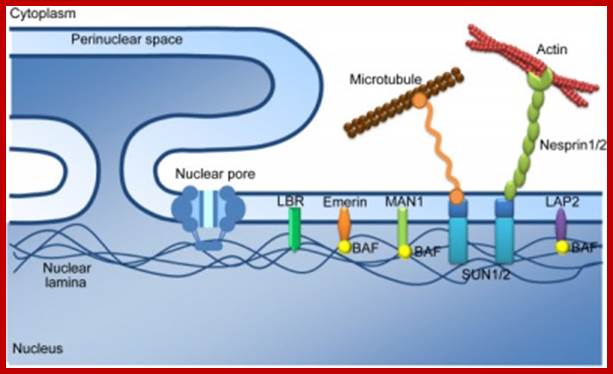
The outer nuclear membrane is associated with microtubules and actin filamentshttps://cellbiology.med.unsw.edu.au
The Nuclear lamina, or the inner nuclear membrane (INM), is a scaffold-like network of protein filaments surrounding the nuclear periphery. This scaffold is made of mostly the type V intermediate filament proteins, lamin A/C and B, which together form a complex meshwork underneath the INM Reviewed in Foisner, 2001; and Wilson et al., 2001) . The lamins are coiled-coil structures that contain a small N-terminal head followed by a rod-like domain (coiled-coil) and a C-terminal globular tail. Via these coiled-coil regions lamins can form parallel dimers, which in turn form polymers with other lamin dimers in an anti-parallel manner (head-to-tail). Although quite resistant to biochemical extraction, the nuclear lamina is nonetheless dynamic and can depolymerisenpd.hgu.mrc.ac.uk
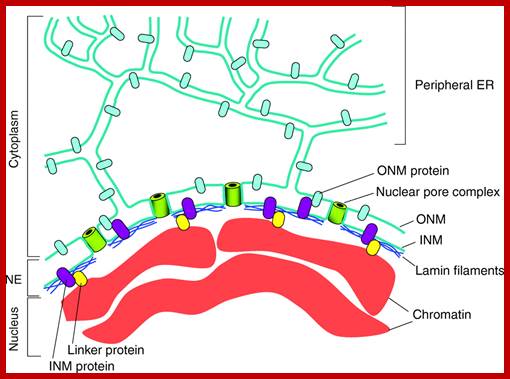
http://jcs.biologists.org/
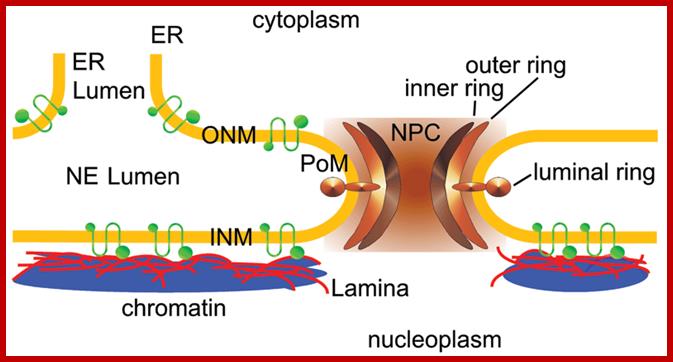
The nuclear envelope: The NE is an integral part of the ER-membrane network (in blue-green). The inner nuclear membrane (INM) and outer nuclear membrane (ONM) connect at sites of NPCs (green barrels) where the membrane curves as it surrounds the NPC. The ONM is continuous with the peripheral ER. The NE contains a variety of proteins that are embedded in the INM (purple) or the ONM (light blue). Most ONM proteins are also found in the peripheral ER. INM proteins can interact with the underlying nuclear lamina (dark blue), with ONM proteins or with chromatin (red), often through linker proteins (yellow). For a detailed description of the various proteins associated with the NE see recent reviews .
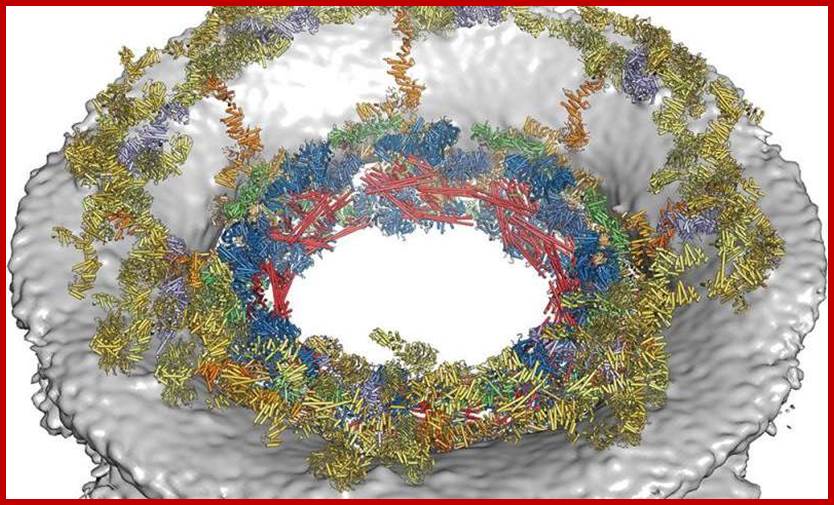
http://reasonandscience.heavenforum.org/
Nucleus is surrounded by a lipid membrane called the nuclear envelope.Until the 1990s, but the nuclear membrane was considered to act as a barrier to protect the chromosome from external factors, from a subsequent study, 1) nuclear membrane is not just a lipid membrane, very complicated structure it is a body (Fig. 1), 2) that the dynamic behavior through the cell cycle, 3) involved in chromosome arrangement, such as carrying out a transcription control, mechanism for controlling the life phenomenon directly has become clear . the order to clarify the association of the structure and function of the nuclear membrane, 1. Mass transport of leaders between the nuclear over cytoplasmic: nuclear pore complex 2. Nuclear membrane collapse and formation process 3. The role of the nuclear membrane in the membrane protein in heterochromatin formation in the nuclear membrane directly under by focusing on the three we are researching.
The structure of the nuclear membrane
nuclear membrane is,
Kakumakugaimaku within the film, it is a complex structure composed of the
nuclear membrane in the membrane-specific protein, nuclear lamina, from a
number of functional sites such as the nuclear pore complex (NPC). Through the SUN proteins of the
nuclear membrane in the membrane protein, it has led the nuclear lamina and
cytoskeleton.
http://reasonandscience.heavenforum.org/
ONM and INM= outer and inner nuclear embranes; www.zotopic.com
NPC side view; http://cronodon.com/BioTech/Cell_Nucleus.html
NPC – side view in section, showing outer and
inner
nuclear membranes (in blue); cytoplasm above,
nucleoplasm below; http://study.com/academy
NPC - view from 'above' (from
cytoplasmic side); http://study.com/academy
NPC - view from 'below' (from nuclear
side; http://study.com/academy
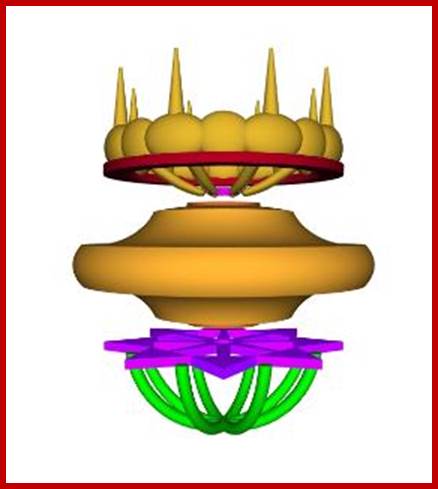
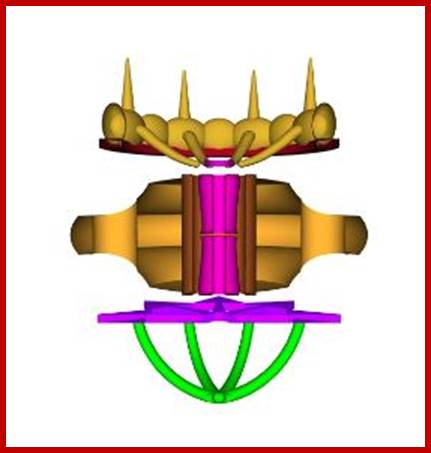
Above four figures are a 3D model of the nuclear
pore complex (NPC).
The NPC is more than a simple molecular sieve,
it has a
mechanism that transports selected materials
into and out
of the nucleus in a regulated manner. The
structure is
made of about 100 proteins (in mammals, about 30
in
yeast) and is about 40 nm in diameter. This
structure is a
remarkable (and complex) molecular nano-machine.
Click
images to enlarge.
Schematic diagram of the nuclear envelope showing the nuclear membranes, nuclear lamina, and pore complexes; Selected integral proteins of the inner nuclear membrane and their topologies are also shown;LBR=laminB receptor; LAP, lamina-associated polypeptide. http://physiologyonline.physiology.org/
Usually, the number of pores present is 40-145 per cm2, but when cells are active, the number of pores increases considerably thousands or more. However the nuclear pores are uniformly distributed with an internuclear space of 150-240 mm.
The nuclear pore is not just a gap, but consists of a complex structural organization. Rockefeller scientists have identified 450-pore proteins At the peripheral region of the pore both the membranes are in continuity. Careful observations reveal the presence of octagonal shaped structures at margin at both the surfaces. Such a structure is called as annulus. In fact, the annulus is made up of 8 globular proteins, which are found at both the surfaces of the nuclear pores. The globular proteins are characteristic and are found in most of the eukaryotic plants and animals as conserved species of the pore is never constant and exhibits expansion and contraction whenever the need arises. Furthermore, the inner surface of the pore complex i.e. towards nuclear sap is associated with chromatin material.
Biogenesis of nuclear envelope:
During cell divisions, the nuclear membrane disappears at the end of prophase but reappears at the end of telophase. Depolymerization of lamins makes the membrane into fragmented vesicles and nuclear pore complexes also separate into vesicular structures. Time-lapse micro photographic studies combined with electron microscopic observations indicate that the nuclear membrane is derived from endoplasmic reticulum. First, vesicles containing pore complexes start associating with chromosomal material, even ER surround the chromosomal clumps and the membranes fuse laterally with each other to form a complete nuclear envelope. Structural organization of the lamins facilitates the biogenesis of nuclear membrane. Microtubules and microtrabaculae play a significant role in the regeneration of nuclear membranes.
Function of Pore Complex:
Nuclear pore is not a passive channel. It does not allow the movement of ions like Na, K, CI, Po4, etc by free diffusion and it acts like a selective sieve. Many nuclear components like DNA polymerase, RNA polymerase, histones, ribo-proteins, etc that are synthesized in cytoplasm readily find their way into nucleus through pore complexes.
http://www.uic.edu/
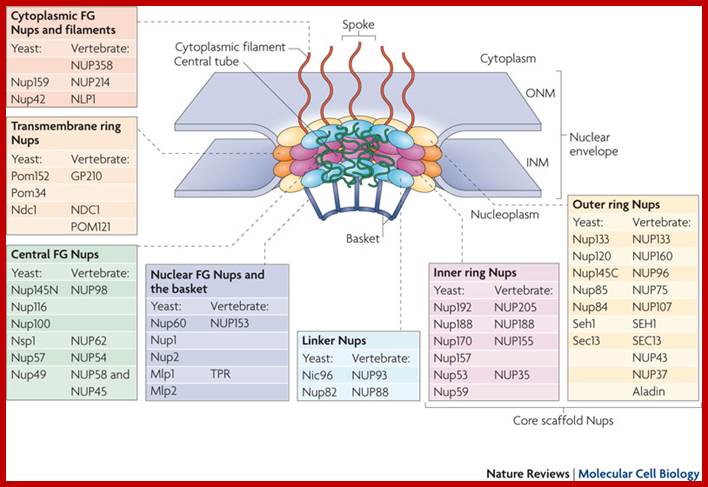
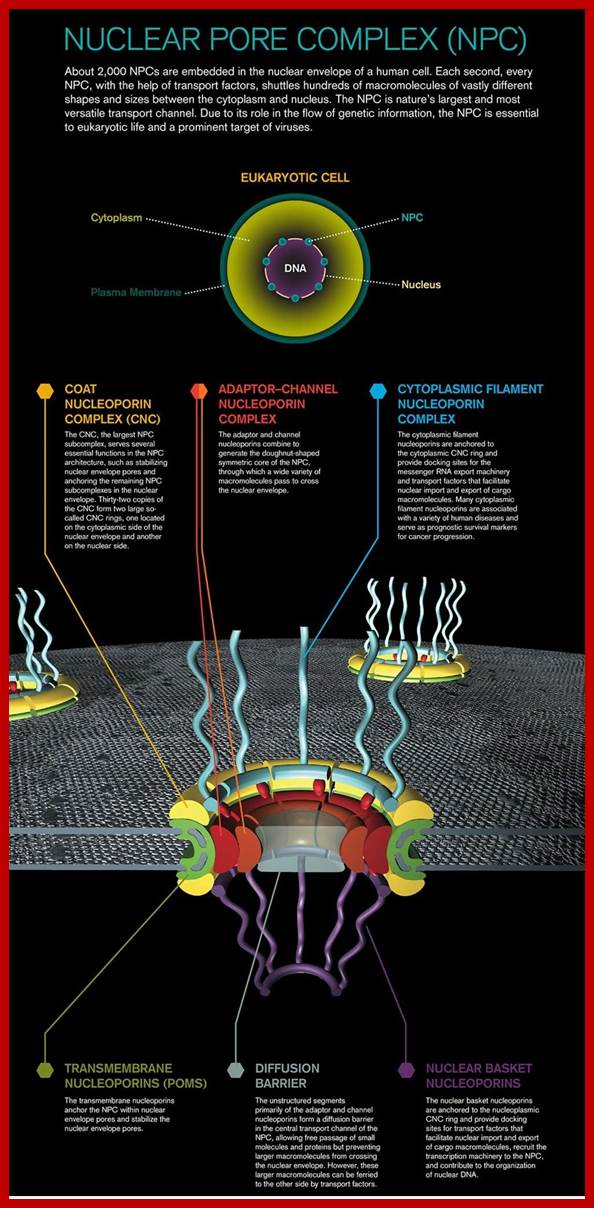
Each nuclear pore complex (NPC) is a cylindrical structure comprised of eight spokes surrounding a central tube that connects the nucleoplasm and cytoplasm. The outer and inner nuclear membranes (ONM and INM, respectively) of the nuclear envelope join to form grommets in which the NPC sits. The NPC is anchored to the nuclear envelope by a transmembrane ring structure that connects to the core scaffold and comprises inner ring and outer ring elements. Linker nucleoporins (Nups) help anchor the Phe-Gly (FG) Nups such that they line and fill the central tube. NPC-associated peripheral structures consist of cytoplasmic filaments, the basket and a distal ring. The Nups that are known to constitute each NPC substructure are listed, with yeast and vertebrate homologues indicated. Both inner and outer ring Nups are known to form biochemically stable NPC subcomplexes, which are thought to have a role in NPC biogenesis and nuclear envelope assembly. GP210, glycoprotein 210; Mlp, myosin-like protein; Ndc1, nuclear division cycle protein 1; Nic96, Nup-interacting component of 76 kDa; NLP1, Nup-like protein 1; Pom, pore membrane protein; Seh1, SEC13 homologue 1; TPR, translocated promoter region.;Nuclear pore complex components www.nature.com

The nuclear pore complex (NPC) associates with numerous molecules and structures in the cytoplasm and nucleoplasm through its cytoplasmic filaments and nuclear basket, respectively. This enables the NPC to be involved in diverse functions in addition to the import and export of soluble and membrane proteins, which requires nuclear transport factors such as karyopherins (Kaps). The basket is part of an interconnected and highly dynamic molecular platform on the nucleoplasmic face of the nuclear envelope. In Saccharomyces cerevisiae and Drosophila melanogaster, this molecular platform couples transcriptional regulation (through interactions between the SAGA chromatin remodelling complex and active genes, interaction of the NPC with components of the TREX2 complex, and interactions between small ubiquitin-related modifier (SUMO) regulatory complexes and the proofreading machinery of exporting ribonucleoproteins (RNPs)105), chromatin stability (through interactions with the transcription-coupled DNA repair machinery)3 and chromosome handling during mitosis (through interactions with spindle checkpoint proteins and the spindle)154. On the other side of the nuclear envelope, cytoplasmic filaments link these processes to the protein synthesis machinery and cytoskeleton. These filaments interact with the Gle1–DEAD box protein 5 (Dbp5) RNA helicase complex to ensure close spatial and temporal coordination between the final phases of messenger RNP export and the initiation of mRNA translation at ribosomes56. Cytoplasmic filaments also interact with cytoskeletal structures to direct traffic in and out of the nucleus to the appropriate cellular 'highways' in the cytoplasm64. The network of protein–protein interactions extending from the NPC also includes integral inner nuclear membrane (INM) proteins such as the yeast establishment of silenced chromatin protein 1 (Esc1)92 and the highly conserved LEM domain167 and SUN domain proteins. SUN domain proteins are thought to link toplasmic microtubules with chromatin through direct interactions with KASH domain proteins in the nuclear envelope lumen172, thus underscoring the existence of an extended communication network spanning the nuclear envelope at the NPC and across both the INM and outer nuclear membrane (ONM). http://www.nature.com/
Molecular Biology of The Cell 4th edition; Nuclear pore basket; http://www.pha.jhu.edu/
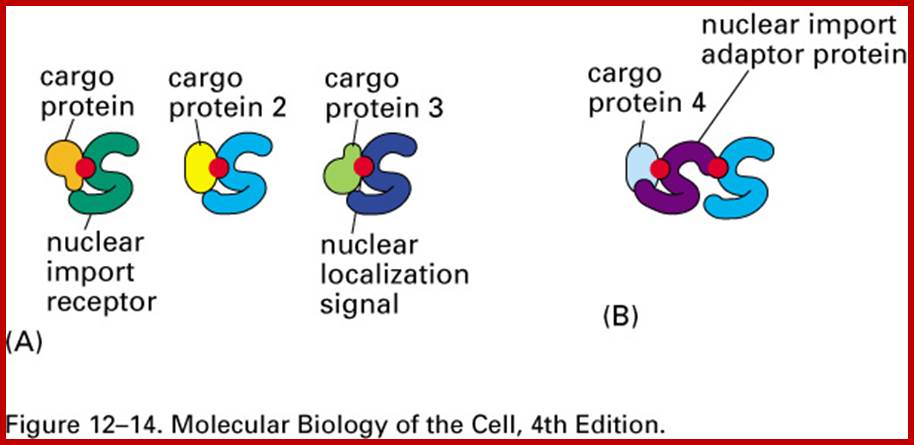
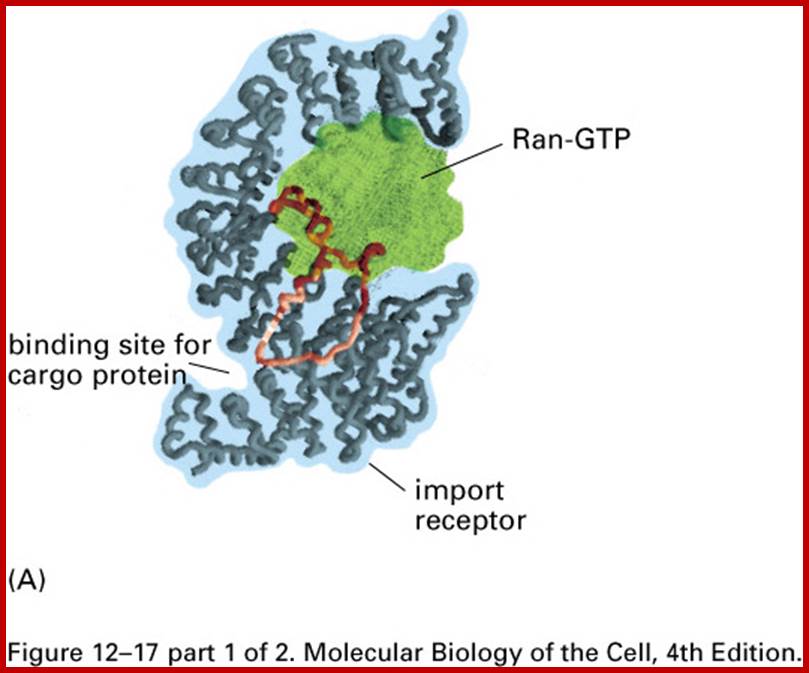
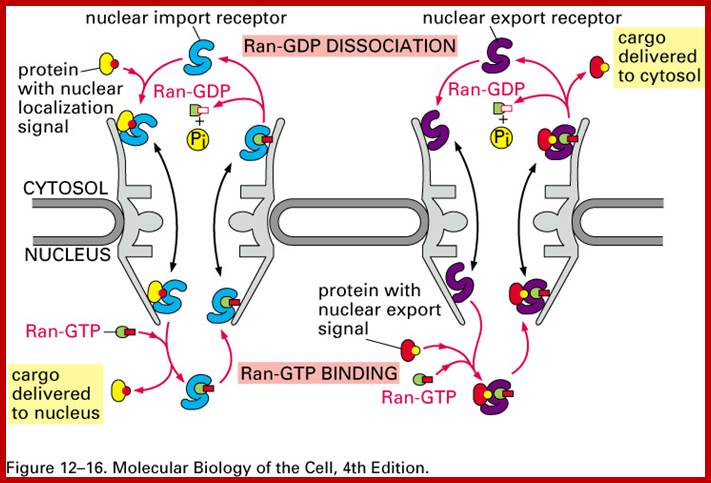
Import and export occur via similar, but non-identical, mechanisms (Figure 12-16 MBC). Molecular cargoes interact with carrier proteins known as importins or exportins (Figure 12-14 MBC; Figure 12-17 MBC). These are members of family of evolutionarily related proteins (the karyopherins). After binding cargo, the importin or exportin has an increased affinity for nuclear pore components, so the karyopherin/cargo complex bound and is translocated across the pore complex; http://www.pha.jhu.edu/
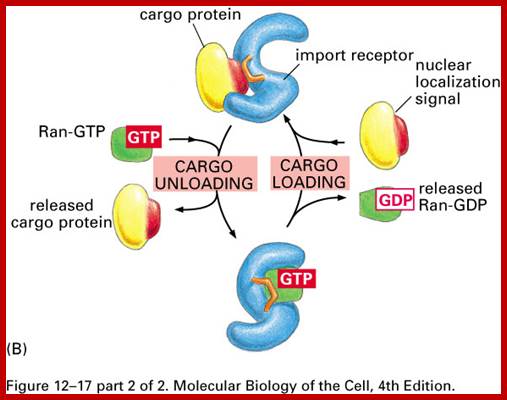
Transport proteins ; http://www.pha.jhu.edu/
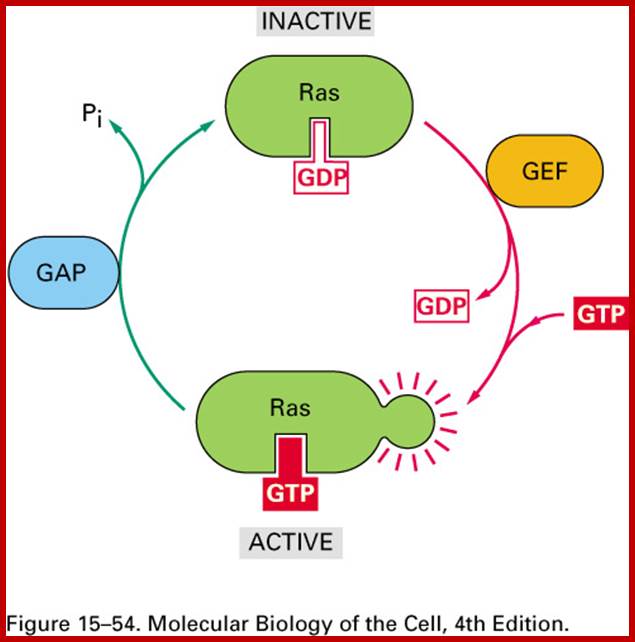
Ras GDP and Ras GTP; http://www.pha.jhu.edu/
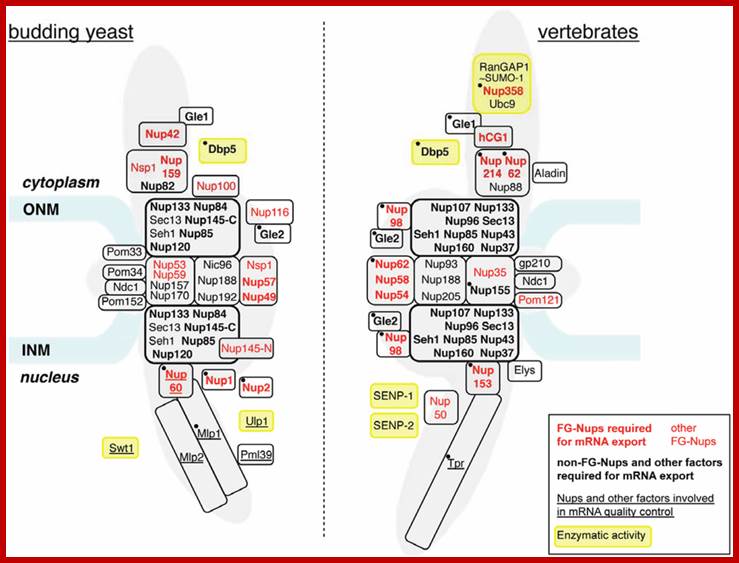
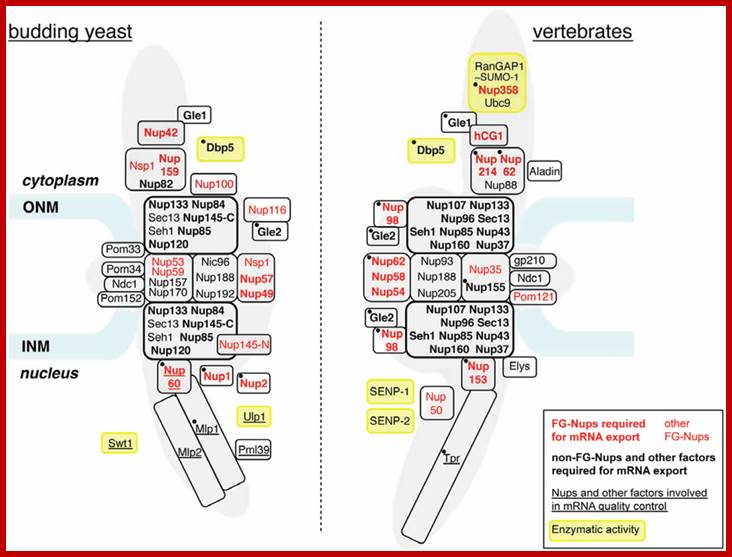
Nucleoporins and nuclear pore complex (NPC)-associated proteins involved in mRNA export. The approximate relative positioning of nucleoporins or NPC subcomplexes within the NPC framework is represented in budding yeast (left) and vertebrates (right) (according to [10]). Phenylalanine-glycine (FG)-nucleoporins appear in red. Nucleoporins (Nups) or NPC-associated proteins with a reported contribution to mRNA export are indicated in bold (seeTable 1). The names of proteins involved in mRNA quality control are underlined (see Table 2). Factors targeted by regulatory events occurring in normal or pathological situations and mentioned in the text (Section 4 and Section 5) are indicated by a black dot. Proteins carrying an enzymatic activity are boxed in yellow. Alternative names for vertebrate nucleoporins are the following: Nup358 = RanBP2; Gle2 = Rae1; Nup35 = Nup53; Nup58 = Nup45; Elys = MEL28; hCG1 = NPL1. The Y-complex is boxed by a thick black line. Note that the inactivation of each Y-complex subunit has not systematically been proven to trigger mRNA export defects: in yeast, seh1 and sec13 mutants do not affect mRNA export [11]; in mammals, mRNA export inhibition has solely been reported upon Nup133 or Nup107 siRNA-mediated depletion [12,13] or upon expression of dominant negative fragments of Nup133 or Nup160 [14]. ONM, outer nuclear membrane; INM, inner nuclear membrane. http://www.mdpi.com/
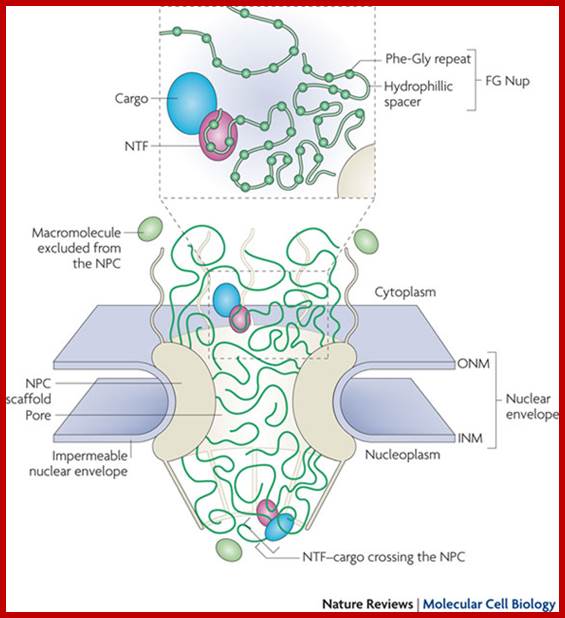
The nuclear pore complex functions as a 'virtual regulated gate'.
The outer and inner nuclear membranes (ONM and INM, respectively) of the nuclear envelope join to form a ring-shaped pore structure where the nuclear pore complex (NPC) forms and resides. At the NPC, the nucleus and cytoplasm are connected by a channel, which is filled with flexible, filamentous Phe-Gly nucleoporins (FG Nups). Spurious macromolecules are physically excluded from entering the densely packed FG Nup meshwork. Nuclear transport factor (NTF)-bound cargo can enter the channel from either its cytoplasmic or nucleoplasmic side and hop between binding sites on the FG Nups until they return to the original compartment or reach the opposite side of the NPC.
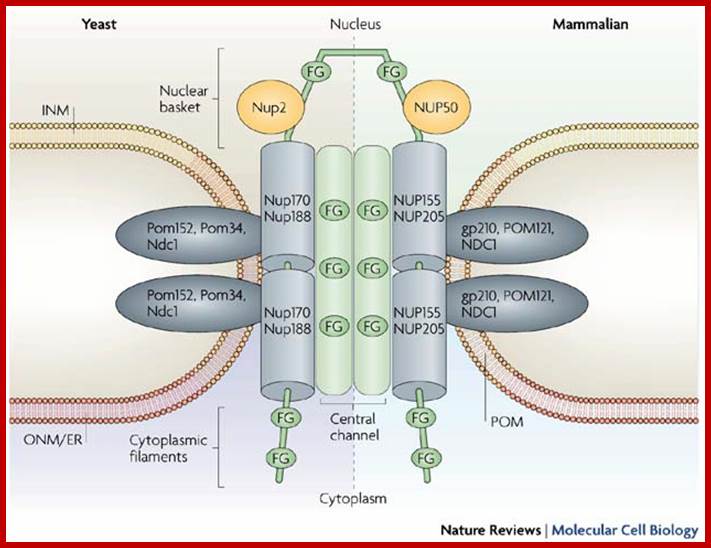
Architecture of yeast (left) and mammalian (right) nuclear pore complexes (NPCs); NPCs mediate the bidirectional exchange of molecules between the cytoplasm and the nucleus. In addition, membrane proteins destined for the inner nuclear membrane (INM) must move from the outer nuclear membrane (ONM) across the pore membrane (POM). The general architecture of the NPC consists of nucleoporins that contain repetitive motifs of Phe-Gly (FG) amino-acid residues (green) that sit on a scaffold of non-FG-nucleoporins (including Nup170 (nucleoporin of 170 kDa) and Nup188 (their mammalian homologues, NUP155 and NUP205, are shown on the right). The scaffold is embedded into the POM by integral POM nucleoporins (Pom152 (POM protein of 152 kDa), Pom34, Ndc1 (nuclear division cycle-1) in yeast and gp210 (glycoprotein of 210 kDa), POM121 and NDC1 in mammals). The FG-nucleoporins are thought to create a continuous surface of FG-repeats that extend from the cytoplasmic filaments through the central channel to the nuclear basket. Nup170, Nup2, Nup188, Pom152 and gp210 have been shown to function in INM protein transport. ER, endoplasmic reticulum. http://www.mdpi.com/
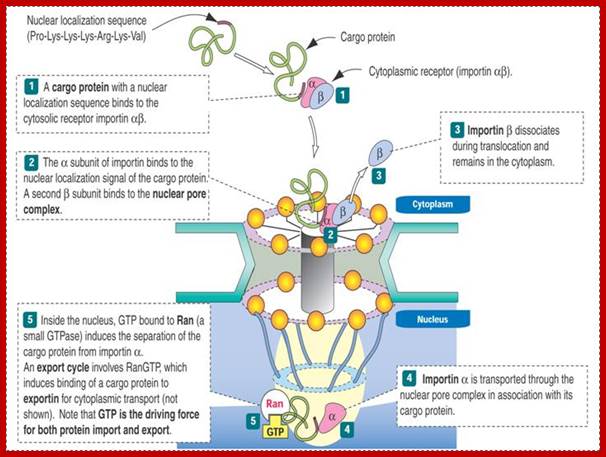
According to the scheme, the protein is recognized by α-importin in complex with β-importin. The α subunit thus recognizes the nuclear localization signal on the protein, and the β subunit is recognized by the pore complex proteins. As the complex begins to be transported by means unknown, the β-subunit is released back into the cytoplasm, and the protein-α subunit complex goes through the pore together. So how do we free the protein? We use a competing protein, Ran-GTP (see #5 above) that displaces the transported protein from the α-subunit without itself binding the carrier. Thus the protein is free to function in the nucleus and the α-subunit then finds its way back into the cytoplasm. But what happens to Ran-GTP? And how do proteins get out of the nucleus and into the cytoplasm? And if they have a chaperone, how do they get loose? The answer involves biological switches. ;classes.kumc.edu.
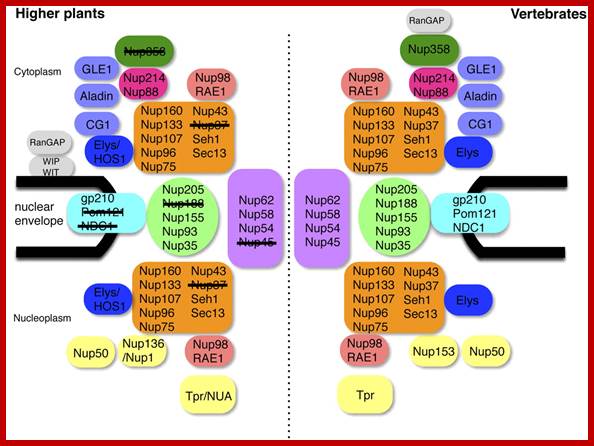
Comparison between the NPCs of Higher Plants and Vertebrates. Subcomplexes are shown as single units. Plant nucleoporins that were not identified in this study are crossed out. WIT, WIP, and RanGAP were identified previously (Xu et al., 2007a; Zhao et al., 2008). http://www.plantcell.org/
Nuclear pore complex is molecular machine acts as a basket;
This massive complex of proteins screens what is required and what is not required, required are transported in and those required in cytoplasm are transported out. This structure has been called ‘holy Grail”On the other hand other materials not required by the nucleus are prevented. Similarly, processed mRNA is transported out but hnRNAs' transportation into cytoplasm is prevented. Ribosomes, processed tRNAs, few sn and sc RNAs, si and miRNAs find their way out into cytoplasm through pore complexes. Some of the snRNAs move out of the nucleus and get modified and return to the nucleus where they have functions. However, the transportation of materials across the pore complex is ATP and GTP dependent and it is a facilitated process. Certain protein components found in the pore complex exhibit ATPase activity.
For reasons not clear one finds packed mRNA into linear transcription when it is transported out. There are at least 30 or proteins that reulated traffic through Pore complex. One of these proteins in pore complex at inner surface magnetically couples with a special molecule- ‘helicase’ that unwinds packed mRNA particles into linear one. During this process most of the mRNA associated proteins complexes as a ball ere unfurled into free mRNA.
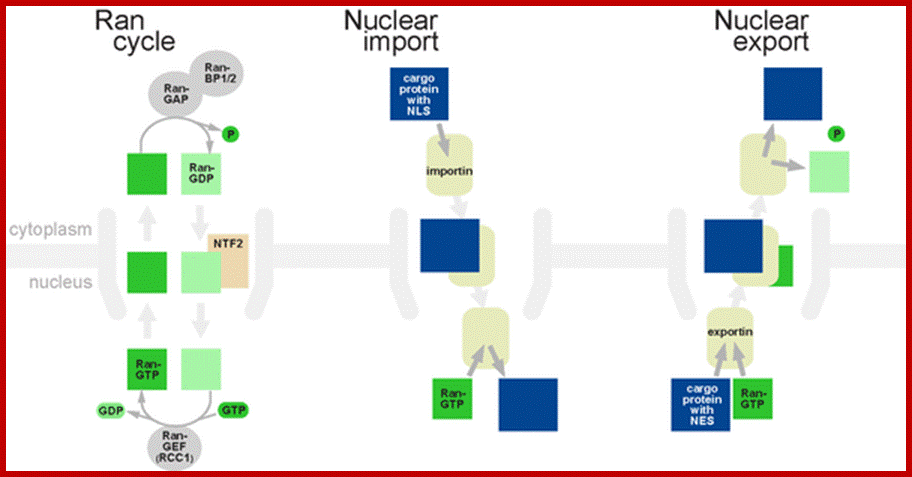
Ran GTP cycle dorim.mokpo.ac.kr
Nuclear import:
Any cargo with a nuclear localization signal (NLS) exposed will be destined for quick and efficient transport through the pore. Several NLS sequences are known, generally containing a conserved phospholipids sequence with basic residues such as PKKKRKV. Any material with an NLS will be taken up by importins to the nucleus.
The classical scheme of NLS-protein importation begins with Importin-α first binding to the NLS sequence, and acts as a bridge for Importin-β to attach. The importinβ—importinα—cargo complex is then directed towards the nuclear pore and diffuses through it. Once the complex is in the nucleus, RanGTP binds to Importin-β and displaces it from the complex. Then the cellular apoptosis susceptibility protein (CAS), an exportin which in the nucleus is bound to RanGTP, displaces Importin-α from the cargo. The NLS-protein is thus free in the nucleoplasm. The Importin-β-RanGTP and Importin-α-CAS-RanGTP complex diffuses back to the cytoplasm where GTPs are hydrolyzed to GDP leading to the release of Importinβ and Importinα which become available for a new NLS-protein import round.
Export of proteins
Some molecules or macromolecular complexes need to be exported from the nucleus to the cytoplasm, as do ribosome subunits and messenger RNAs. Thus there is an export mechanism similar to the import mechanism.
In the classical export scheme, proteins with a nuclear export sequence (NES) can bind in the nucleus to form a heterotrimeric complex with an exportin and RanGTP (for example the exportin CRM1). The complex can then diffuse to the cytoplasm where GTP is hydrolyzed and the NES-protein is released. CRM1-RanGDP diffuses back to the nucleus where GDP is exchanged to GTP by RanGEFs. This process is also energy dependent as it consumes one GTP. Export with the exportin CRM1 can be inhibited by Leptomycin B
Export of RNA;
There are different export pathways through the NPC for each RNA class that exists. RNA export is also signal mediated (NES); the NES is in RNA-binding proteins (except for tRNA which has no adapter). It is notable that all viral RNAs and cellular RNAs (tRNA, rRNA, U snRNA, microRNA) except mRNA are dependent on RanGTP. Conserved mRNA export factors are necessary for mRNA nuclear export. Export factors are Mex67/Tap (large subunit) and Mtr2/p15 (small subunit). In higher eukaryotes, mRNA export is thought to be dependent on splicing which in turn recruits a protein complex, TREX, to spliced messages. TREX functions as an adapter for TAP, which is a very poor RNA binding protein. However, there are alternative mRNA export pathways that do not rely on splicing for specialized messages such as histones. Recent work also suggest an interplay between splicing-dependent export and one of these alternative mRNA export pathways for secretory and mitochondrial transcripts. http://en.wikipedia.org/
Nuclear Sap:
The amorphous liquid found within the nucleus is often referred to as karyolymph. Chromatin material, nucleoli and other enzymatic components are suspended in this liquid. In addition, the fluid at the inner nuclear membrane is pervaded with another network of fibrils, which occupy the entire cortical region of the nuclear sap. The fibrils are lamins, they are a kind of intermediate filaments. The nuclear sap also contains another kind of network of proteins, unique to the nuclear sap called nuclear matrix a heterogeneous kind of proteins. Within such a network, chromatin material, various species of RNAs and enzyme components are found embedded. The exact role of such a network of is not known.
Apart from the network, the fluid has the enzymatic components required for chromosomal duplication, transcription and processing and repair activities. During meiosis, specialized structures like synaptonemal complex, appearance at zygotene stage and disappearance at diplotene stage, very dramatic.. Nuclear sap also posses certain enzymatic activities like kinases, methylases, acetylases and few others, but respiratory activities like glycolysis, is totally absent.
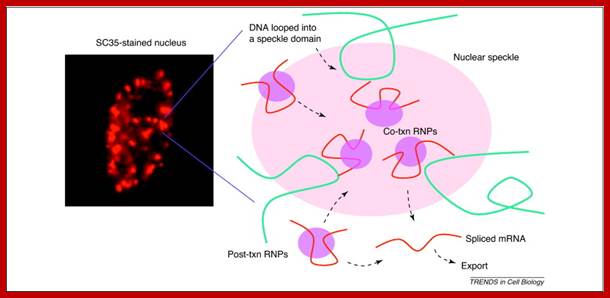
4. Nuclear Speckles:
Speckles are irregular shaped structures of varied size and the nucleus
typically contains 25-50 of these sub-nuclear bodies (Spector 2001; Lamond and
Sleeman 2003). Nuclear speckles are rich in splicing factors including small
nuclear ribonucleoprotein particles (snRNPs) and non-snRNP protein splicing
factors such as the splicing factor SC35. Speckles are often found
close to actively transcribed genes and are thought to act as a reservoir for
the splicing of nascent pre-mRNA at nearby genes.
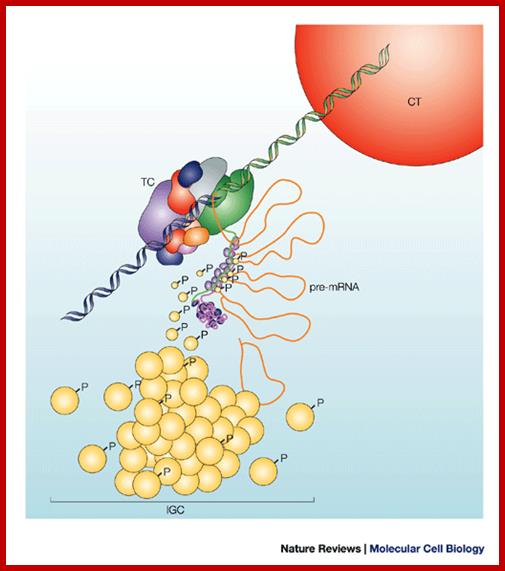
Pre-mRNA splicing; Joon hee Han et al; http://www.sciencedirect.com/
Nuclear speckles are interchromatin granules are dynamic; clusters of proteins associated with pre-mRNAs and they perform splicing ; http://www.nature.com/
5. Cajal Bodies:
Numerous Cajal bodies, or coiled bodies are found in many cell types and are typically 0.2-1 um in diameter (Matera 1999). These structures appear as a tangle of coiled threads and are characterized by the presence of the p80 coilin protein. Cajal bodies are thought to play a role in snRNP biogenesis and in the trafficking of snRNPs and small nucleolar RNPs (snoRNPs). Cajal bodies are rich in spliceosomal U1, U2, U4/U6 and U5 snRNPs as well as U7 snRNP involved in histone 3’-end processing (most histone transcripts are not polyadenylated rather their 3’ ends are produced by an endonucleolytic cleavage) and U3 and U8 snoRNPs involved in processing of pre-rRNA. It is believed that snRNPs and snoRNPs move through Cajal bodies then on to nuclear speckles or nucleoli respectively.
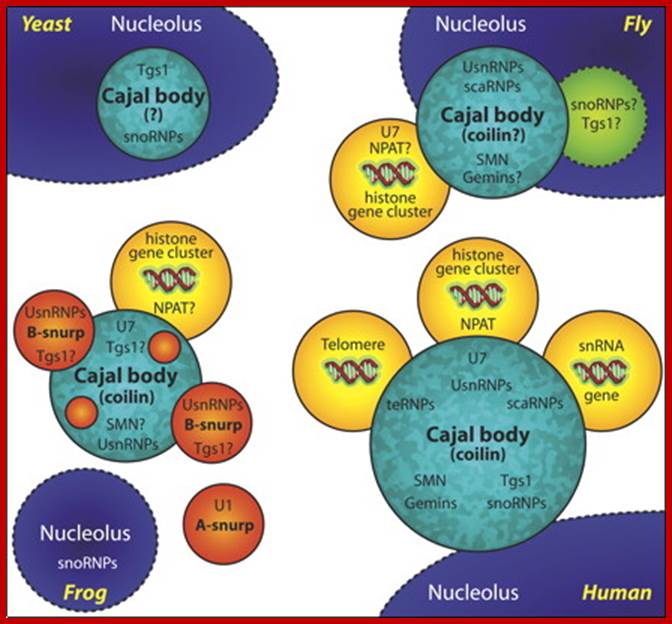
A schematic of Cajal bodies and their relationship to nucleoli in yeast, fly, frog, and human: The various components that are known to accumulate in these structures are shown; question marks denote those that have not be experimentally demonstrated. The yeast nucleolar body (herein referred to as a Cajal body) is the least well studied of the four. Only two endogenous factors are known to accumulate in this structure, snoRNPs and the trimethylguanosine synthase protein, Tgs1. Interestingly, when human SMN is ectopically expressed (budding yeast do not encode a recognizable homologue) it accumulates in the Cajal body. Amphibian oocyte nucleoli are extrachromosomal. In the oocyte germinal vesicle, the Cajal body has a modular structure composed of B-type “snurposomes” and a Cajal body matrix that contains, among many other factors, coilin and the U7 snRNP. B-type inclusions are shown within the matrix of the frog Cajal body; A-type snurposomes are thought to accumulate only U1 snRNPs. Somatic Cajal bodies have not been well studied in the frog. A conserved feature among Cajal bodies in humans, flies, and frogs is a frequent association with the cell cycle–dependent histone gene clusters. Human Cajal bodies additionally associate with snRNA genes and telomeres. Moreover, a class of small Cajal body–specific RNPs (scaRNPs), including telomerase, accumulate in human Cajal bodies. Thus, human Cajal bodies appear to have more accessories hanging in and around them than do the respective structures in other eukaryotes. Now that the Cajal body has been identified in the fly, a clearer evolutionary picture is sure to emerge (JCB).
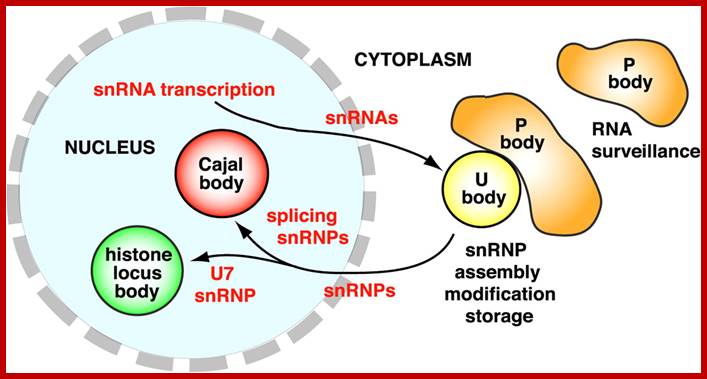
Ji-Long Liu and Joseph G. Gall ; http://www.pnas.org/
U bodies contain snRNPS; several of them like U2 and U7 and U85 scaRNA; some of them are specific to cajal bodies; they are involved in mRNA processing. Cytoplasmic U bodies associate with P bodies, which function in RNA surveillance and decay.
Processing bodies called P bodies are involved in mRNA turnover, they are found in plant and animal cells. Processing bodies (P-bodies) are distinct foci within the cytoplasm of the eukaryotic cell consisting of many enzymes involved inmRNA turnover. P-bodies have been observed in somatic cells originating from vertebrates and invertebrates, plants and yeast. To date, P-bodies have been demonstrated to play fundamental roles in general mRNA decay, nonsense-mediated mRNA decay,adenylate-uridylate-rich element mediated mRNA decay, and microRNA induced mRNA silencing.[1] Not all mRNAs which enter P-bodies are degraded, as it has been demonstrated that some mRNAs can exit P-bodies and re-initiate translation.[2][3]
The following activities have been demonstrated to occur in or to be associated with P-bodies: decapping and degradation of unwanted mRNAs; storing mRNA until needed for translation; aiding in translational repression by miRNAs (related to siRNAs). Proteins involved in mRNA silencing by miRNA require P bodies.
6. Gems (Gemini of Cajal Bodies);
Gems are found adjacent to Cajal bodies and are characterized by the presence of the survival of motor neurons gene product (SMN) and Gemin 2 (Spector 2001; Lamond and Sleeman 2003). Cytoplasmic SMN and Gemin 2 are involved in the assembly of snRNPsand therefore the nuclear pool may play a role in snRNP maturation. Spinal muscular atrophy, a motor neuron disorder, results from reduced levels or a mutation in SMN proteins.
7. Promyelocytic Leukemia (PML) nuclear Bodies
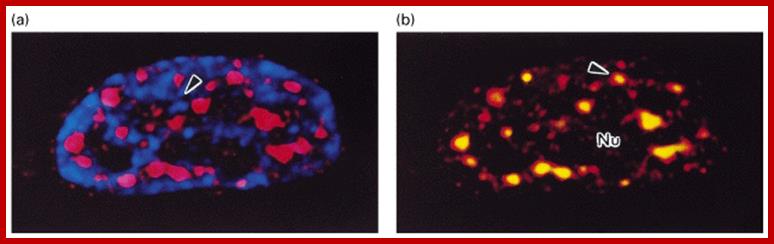
PML bodies are characterized by the presence of PML protein (Strouboulis and Wolffe 1996; Spector 2001; Lamond and Sleeman 2003). PML bodies vary in size from 0.3-1 um in diameter and a nucleus typically contains 10-30 of these structures (Figure 2). The primary role of PML bodies remains unclear; but they may play a role in transcriptional regulation and in anti-viral responses. Individuals suffering from acute Promyelocytic leukemia (APL) have a translocation in which the PML gene is fused to the gene encoding the alpha-retinoic acid receptor, resulting in the production of a fusion protein. Cells from these individuals exhibit fragmented PML bodies. However, treatment of APL patients with retinoic acid results in cancer remission and the restoration of normal PML body structure. http://www.abcam.com/
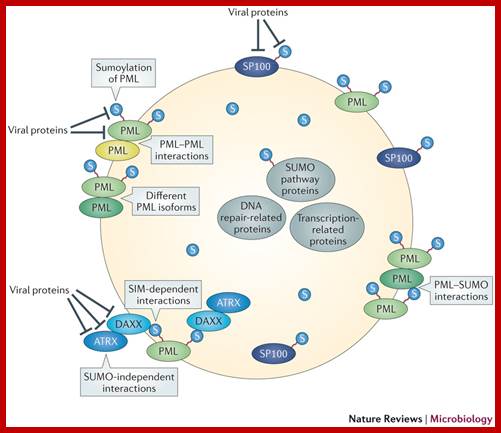
Roger D. Everett et al, ;http://www.nature.com/
PML nuclear body (PML NB) assembly requires (shown in grey boxes): small ubiquitin-like modifier (SUMO; denoted S in the figure) modification of promyelocytic leukaemia protein (PML); PML self-interaction through its coiled-coil structure in its tripartite motif (TRIM); the presence of different PML isoforms (no single PML isoform is sufficient to establish fully normal structures120); SUMO–SUMO interaction motif (SIM)-dependent interactions (such as that between the DAXX SIM and the SUMO attached to PML118, 121); SUMO-independent interactions (such as that between DAXX and ATRX86) (reviewed in Refs 12, 14); and PML–SUMO interactions through the PML SIM115. The residence time of PML NB components in these structures varies from a few minutes to a few seconds, indicating dynamic exchange with the nucleoplasm122, and sumoylation events themselves are likely to be highly dynamic. Several avenues are available for viral proteins to disrupt or modify PML NBs by targeting the aforementioned processes or the proteins involved in PML NB formation. Viral proteins — such as herpes simplex virus type 1 (HSV-1) ICP0 (Refs 67, 68, 69, 123), varicella zoster virus (VZV) orf61 (Refs 72, 73), human cytomegalovirus (HCMV) IE1 (Refs 23, 69), Epstein–Barr virus (EBV) BZLF1 (Ref. 38) and EBNA-1 (Ref. 124), murine herpesvirus 68 (MHV68) orf75c125, Kaposi's sarcoma herpesvirus (KSHV) LANA2 (Ref. 40) and adenovirus 5 E4orf3 (Refs 75, 76) — can disrupt PML NBs by targeting PML or sumoylated PML. Other viral proteins — including HSV-1 ICP0 (Refs 69, 71, 123), BHV-1 BICP0 (Ref. 70), VZV orf61 (Ref. 73), HCMV IE1 (Refs 24, 26), herpesvirus saimiri orf3 (Ref. 126) and EBV EBNA5 (Ref. 127) — have been shown to target SP100 and sumoylated SP100. HSV-1 ICP0, BHV-1 BICP0, EHV-1 EICP0 and pseudorabies virus EP0 reduce the general level of sumoylated proteins in the cell67, 70. This is not a comprehensive list of all the viral proteins that disrupt or interact with PML NBs, or interact or interfere with PML NB proteins (notably DAXX, ATRX and the DAXX–ATRX complex) by mechanisms that are not known to be related to SUMO. (reviewed elsewhere).
Chromosomes are organized into large domains called chromosome territories (Spector 2001; Lamond and Sleeman 2003). Chromosomes exhibit different levels of compaction with the most condensed regions referred to as heterochromatin, which is either lacking in genes or contains genes that are transcriptionally repressed. Less condensed regions of chromosomes are known as euchromatin, which is rich in gene concentration and is usually, but not always, actively transcribed.
Chromatin:
The network of fibers that take up stain in the presence of aceto-orcien, basic fuchsin is called chromatin network. At the interphase stage chromosomes are relaxes and diffused. Such chromosomes appear as a network of fine chromosomal fibers. But with the ones of cell division, such diffused chromosomes undergo a spiral coiling and condense into cytologically visible compact threads. It is at this stage the number of chromosomes can be counted and its morphological features can be made out.
Metaphasic chromosomes are made up of two sister chromatids or chromonemata, which are relationally coiled to each other. At a specific region the double stranded chromonemata appear as if they are held together. In fact this region looks like a non-stainable gap or constriction called centromere. On either side of the centromere a cup shaped protein structures are found. They are called kinetochores. Mitotic chromosomes possess two kinetochores and meiotic chromosomes contain only one kinetochore in the first meiosis, but two appear in the second meiosis. Tractile fibers, which develop during the formation of mitotic apparatus, actually develop from the kinetochore structures. Some of the chromosomes also contain another constriction towards the end of the chromonemata, it is called secondary constriction towards the end of the chromonemata, and it is called secondary constriction or nuclear organizer. However, most of the chromosome contain, minute granular structures called telomeres at their extreme ends.
Chromosomes when viewed through optical microscope appear as highly coiled double stranded threads. But careful analysis of whole mount chromosomes under electron microscopy shows that each chromonema are made up of basic chromonemal fiber of 30 mm thick and several centimeters long. But these fibers are highly coiled into compact structures, which appear cytological as condensed chromosomal strands. The number of chromosomes present in a nucleus is denoted by the terms, like haploid, diploid, etc. The haploid term indicates on set of chromosomes, in which each of the chromosomes is morphologically and genetically distinct and characteristic. The number of chromosomes that constitute a haploid set varies from species to species, but for a given species it is always constant and characteristic. A diploid set is made up of two sets of haploid chromosomes. The multiple sets are denoted as polyploids. Chromosomes are thus considered as vehicles of heredity and act as the repository for genetic information required for an organism.
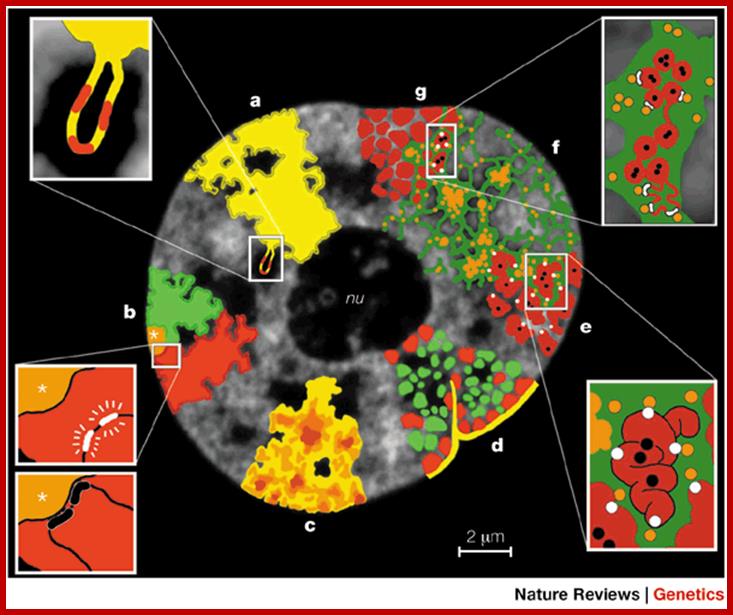
Structural features that support the
chromosome-territory–interchromatin-compartment (CT–IC) model are shown. These
features are drawn roughly to scale on an optical section taken from the
nucleus of a living HeLa cell. Although experimental evidence is available to
support these features, the overall model of functional nuclear architecture is
speculative (see text). a | CTs
have complex folded surfaces. Inset: topological model of gene regulation. A
giant chromatin loop with several active genes (red) expands from the CT
surface into the IC space. b | CTs
contain separate arm domains for the short (p) and long chromosome arms (q),
and a centromeric domain (asterisks). Inset: topological model of gene
regulation. Top, actively transcribed genes (white) are located on a chromatin
loop that is remote from centromeric heterochromatin. Bottom, recruitment of
the same genes (black) to the centromeric heterochromatin leads to their
silencing. c | CTs have variable chromatin density
(dark brown, high density; light yellow, low density). Loose chromatin expands
into the IC, whereas the most dense chromatin is remote from the IC. d | CT showing early-replicating
chromatin domains (green) and mid-to-late-replicating chromatin domains (red).
Each domain comprises ![]() 1 Mb. Gene-poor chromatin (red), is preferentially
located at the nuclear periphery and in close contact with the nuclear lamina
(yellow), as well as with infoldings of the lamina and around the nucleolus
(nu). Gene-rich chromatin (green) is located between the gene-poor
compartments. e | Higher-order chromatin structures
built up from a hierarchy of chromatin fibres. Inset: this topological view of
gene regulationindicates that active genes (white dots) are at the surface of
convoluted chromatin fibres. Silenced genes (black dots) may be located towards
the interior of the chromatin structure. f | The
CT–IC model predicts that the IC (green) contains complexes (orange dots) and
larger non-chromatin domains (aggregations of orange dots) for transcription,
splicing, DNA replication and repair. g | CT with
1 Mb. Gene-poor chromatin (red), is preferentially
located at the nuclear periphery and in close contact with the nuclear lamina
(yellow), as well as with infoldings of the lamina and around the nucleolus
(nu). Gene-rich chromatin (green) is located between the gene-poor
compartments. e | Higher-order chromatin structures
built up from a hierarchy of chromatin fibres. Inset: this topological view of
gene regulationindicates that active genes (white dots) are at the surface of
convoluted chromatin fibres. Silenced genes (black dots) may be located towards
the interior of the chromatin structure. f | The
CT–IC model predicts that the IC (green) contains complexes (orange dots) and
larger non-chromatin domains (aggregations of orange dots) for transcription,
splicing, DNA replication and repair. g | CT with ![]() 1-Mb chromatin domains (red) and
IC (green) expanding between these domains. Inset: the topological
relationships between the IC, and active and inactive genes72. The finest branches of the IC end
between
1-Mb chromatin domains (red) and
IC (green) expanding between these domains. Inset: the topological
relationships between the IC, and active and inactive genes72. The finest branches of the IC end
between ![]() 100-kb chromatin domains. Top: active genes
(white dots) are located at the surface of these domains, whereas silenced
genes (black dots) are located in the interior. Bottom: alternatively, closed
100-kb chromatin domains. Top: active genes
(white dots) are located at the surface of these domains, whereas silenced
genes (black dots) are located in the interior. Bottom: alternatively, closed ![]() 100-kb chromatin domains with
silenced genes are transformed into an open configuration before
transcriptional activation.; Functional Nuclear Architecture- http://www.nature.com/
100-kb chromatin domains with
silenced genes are transformed into an open configuration before
transcriptional activation.; Functional Nuclear Architecture- http://www.nature.com/
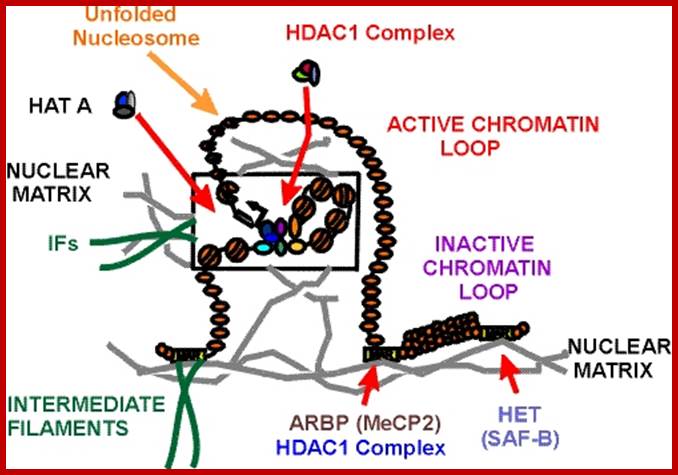

MeCP2 Binds to 5hmC Enriched within Active Genes and Accessible Chromatin in the Nervous System; The high level of 5-hydroxymethylcytosine (5hmC) present in neuronal genomes suggests that mechanisms interpreting 5hmC in the CNS may differ from those present in embryonic stem cells. Here, we present quantitative, genome-wide analysis of 5hmC, 5-methylcytosine (5mC), and gene expression in differentiated CNS cell types in vivo. We report that 5hmC is enriched in active genes and that, surprisingly, strong depletion of 5mC is observed over these regions. The contribution of these epigenetic marks to gene expression depends critically on cell type. We identify methyl-CpG-binding protein 2 (MeCP2) as the major 5hmC-binding protein in the brain and demonstrate that MeCP2 binds 5hmC- and 5mC-containing DNA with similar high affinities. The Rett-syndrome-causing mutation R133C preferentially inhibits 5hmC binding. These findings support a model in which 5hmC and MeCP2 constitute a cell-specific epigenetic mechanism for regulation of chromatin structure and gene expression; http://www.cell.com/
Suspended within the nuclear sap are the net work of threads, when stained they take color, hence they are called chromatin threads (chroma-colour, tene-thread). Some of the threads particularly the ends are associated with either pore complexes or inner nuclear membranes. This interphase chromatin network is not a constant feature, but changes as and when the cell passes through various stages of cell division. Nevertheless, the chromatin at the GI stage, appears to be diffused, thin, single stranded and coiled, but enmeshed in the stranded chromatin threads undergo duplication to form double stranded chromatin threads. During duplication chromosomal DNA replicates, and necessary histones and nonhistones are drawn from the nuclear sap to form sister chromatin threads. With the progress of interphase into prophase, the long, thin threads undergo a continuous process of spiralization, which results in the condensation of long and thin threads into shorter and thicker structures. At the same time, the chromatin distangles from the network and chromosomes slowly get resolved into individual threads.
Chromosome is a threadlike linear strand of DNA and associated proteins in the nucleus of animal and plant cells that carries the genes and functions in the transmission of hereditary information. It is also a circular strand of DNA in bacteria and cyanobacteria that contains the hereditary information necessary for cell life. http://www.imvu.com/
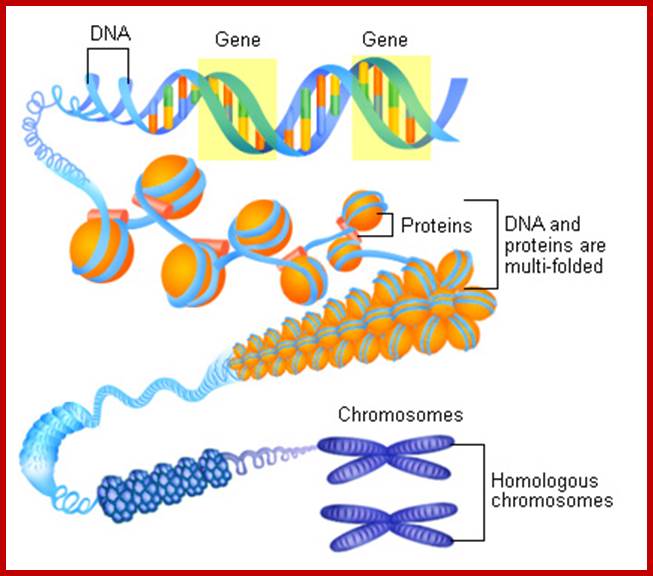
http://activity.ntsec.gov.tw/
By the time, cells reach metaphase stage, chromosomes undergo maximum condensation, and individual chromosomes can be made out. At the stage the number and the detailed structure of them can be studied with light microscope or electron microscope.
Number of chromosomes: The number of chromosomes varies from organism to organism (2-1600) and this number is constant and characteristic for a given species. (only few of are given below in the Table below).
|
Common name |
Specific name |
Chromosomal number(2n) |
Fruity fly |
Drosophila |
8 |
|
Frog |
Rana pipiens |
20 |
|
Gorilla |
Gorilla gorilla |
48 |
|
Pan triglodytes Monkey |
Chimpanzee Macaca mulatta |
48 42 |
|
Man |
Homo sapien sapiens |
46 |
|
Garden pea |
Pisum sativum |
14 |
|
French bean |
Phaseolus vulgaris |
14 |
|
Onion |
Allium cepa |
16 |
|
Cabbage |
Brassica oleracea |
18 |
|
Coffee |
Coffea Arabica |
44 |
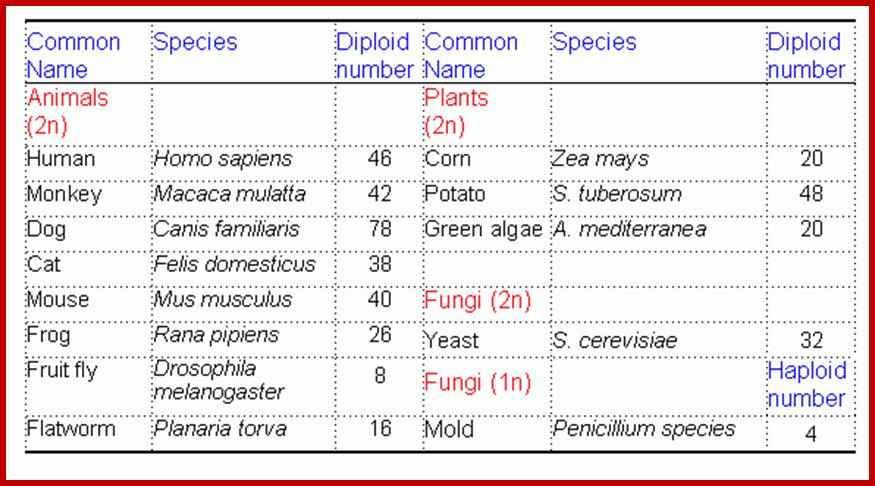
The number of chromosomes is denoted by the terms Karyotype which may be either haploid or polyploid. The haploid karyotype consists of one set of chromosomes, where every individual chromosome is structurally and genomically different from the others and exhibit unique characteristics. For example: in the case of onion, the haploid (n) chromosome number is 8 and let us call them as A, B, C, D, E, F, G, H. Here, each chromosome is different and unique in its genomic content. And such a set of chromosomes is called haploid set. If two such sets of chromosomes are put together in the same nucleus then that nucleus or the organism that posses it, is called diploid i.e. here two haploid sets are present, where A to H chromosomes are present in all respects are called homologous chromosomes or homologous pairs. On the other hand, A and B chromosomes are being different, called non-homologous chromosomes. The terms triploid (3 n), tetraploid (4 n) and polyploid just indicates the number of sets present in the nucleus.
A group of organisms belonging to a particular species though show a constant chromosomal number, say diploid, they often exhibit variation in chromosome numbers, either by loss or gain of one or more chromosomes. In some cases the entire set of chromosomes may be involved. This variation in chromosomal number leads to variation in the morphology and functional behavior of organisms. Such changes may ultimately lead to variation and origin o species. This is one of the fundamental steps in organic evolution.
X and Y chromosomes; www.sciencephoto.com
Size of chromosomes: Chromosomal size varies from organism to organism, however a particular size of chromosomes is constant for a given species. Some of the plants of cyperaceae and Luzyla have very small chromosomes, but plants like Trillium have quite large chromosomes of the size 30µ in length. However, in a given karyotype, all the chromosomes are not of the same size (asymmetrical karyo-type) and rarely do we find organisms with chromosomes of the same size (symmetrical karyotype). Some chromosomes like salivary gland chromosomes (Drosophila), lampbrush chromosomes, (Xenopus levis) and chromosomes in the endosperm haustoria of Phaseolus, are 100-1000 times larger than their somatic chromosomes. These are called special type of chromosomes or giant chromosomes.
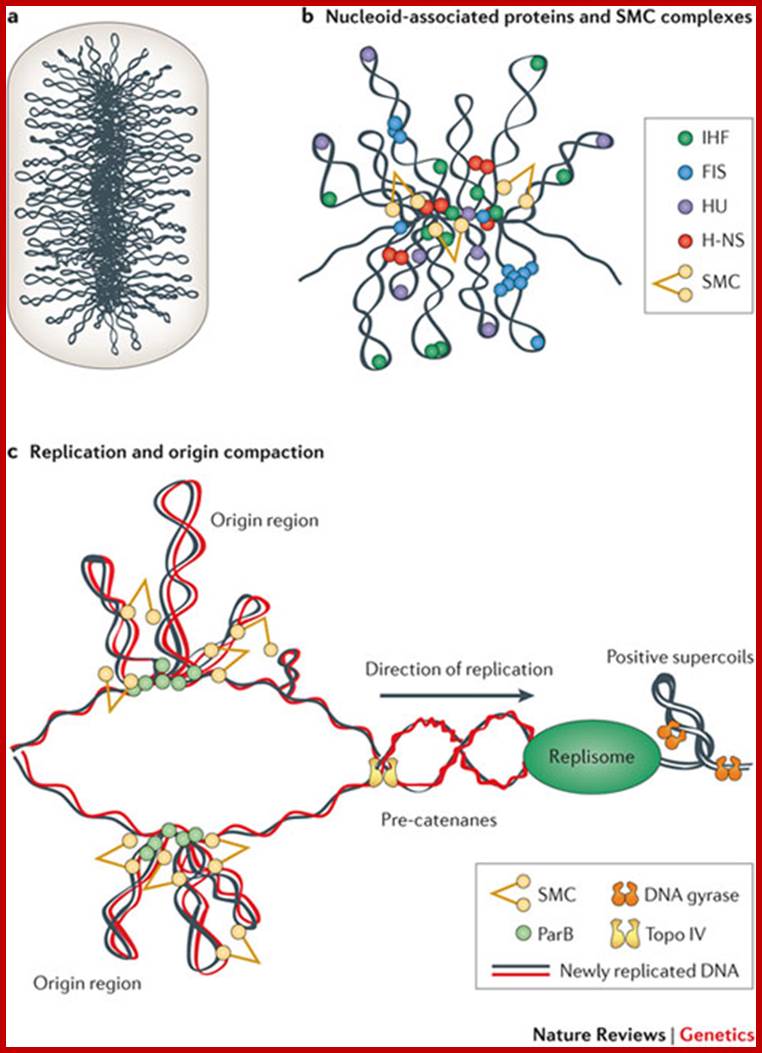
Organization and Segregation of bacterial Chromatin; Bacterial DNA equies 1000 fold compaction to fit into bacterial space. Xindan Wang et al;
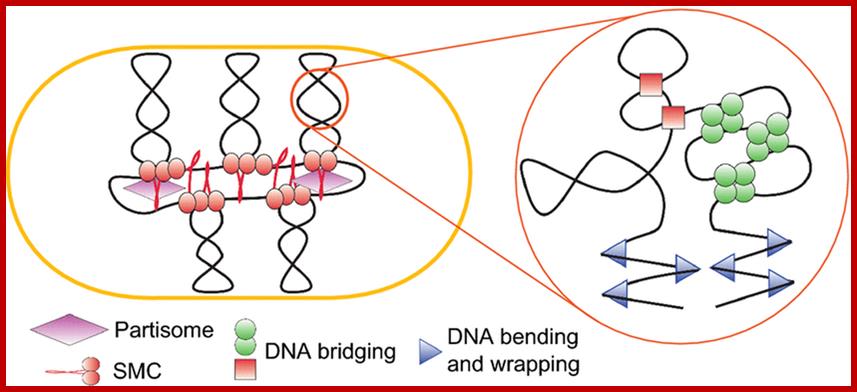 Bacterial chromosome (DNA with bound specific
proteins):. Chromosome structure is stabilized by various NAPs that bend and
bridge DNA, supercoiling, which energizes and compacts DNA, and condensins,
which stabilize giant loops and tether them to extra chromosomal elements; http://femsle.oxfordjournals.org/
Bacterial chromosome (DNA with bound specific
proteins):. Chromosome structure is stabilized by various NAPs that bend and
bridge DNA, supercoiling, which energizes and compacts DNA, and condensins,
which stabilize giant loops and tether them to extra chromosomal elements; http://femsle.oxfordjournals.org/
Picturing the organization of mitotic chromosomes; R.Mark Wilson;
A new study simulates the unpacking and repacking of a human chromosome during cell division.
Chromosomes, with their tightly coiled but elongated X-shaped profile, are among the most recognizable features of a dividing cell. But how DNA is organized inside the chromosomes is largely unresolved. Although a chromosome may look hopelessly tangled, there exist distinct patterns in how it contorts and arranges itself. Methods known collectively as chromosome conformation capture, first developed by Job Dekker of the University of Massachusetts Medical School and colleagues a decade ago, offer a way to examine the folded conformations by chemically linking parts of the chromosome that are spatially close. Sequencing the linked DNA segments allowed the researchers to figure out which parts of the chromosome are likely to have intersected. The result was a map of contact points at which segments of base pairs fold inside a nucleus (see Physics Today, December 2009, page 19). Dekker, Leonid Mirny (MIT), and their colleagues have now combined chromosome conformation capture with polymer simulations of DNA to model how a chromosome disassembles and then reassembles itself during mitosis. They found that as the cell nucleus is dissolved, chromosomes become reorganized in two phases. First, the long chromosome fiber of protein and DNA compacts itself into an array of consecutive loops of some 80 000 to 120 000 base pairs that emanate from and return to a central scaffold—the flexible, dark-colored rod pictured here. That phase is then followed by axial compression of the scaffold to form a short, dense cylinder. This movie by the Mirny lab illustrates the process. Yet to be understood is what interactions guide the reorganization. (N. Naumova et al., Science, 342, 948, 2013.); R. Mark Wilson; http://scitation.aip.org/—
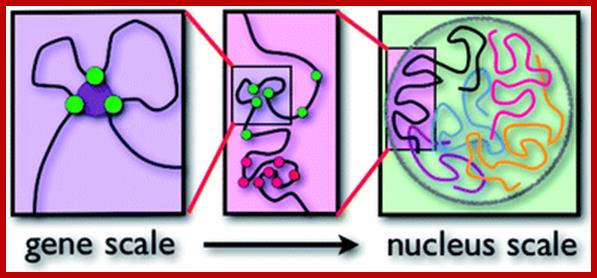
Polymer physics, scaling and heterogeneity in the spatial organization of chromosomes in the cell nucleus; M. Barbieri et al; http://pubs.rsc.org/
We the authors review some of the recent progresses made by the application of polymer physics to the understanding of the spatial organization of chromosomes in the cell nucleus. In particular, we focus on some scaling properties of the contact probability of pairs of sites, discovered by recent genome-wide experiments exploiting new technologies such as Hi-C methods.
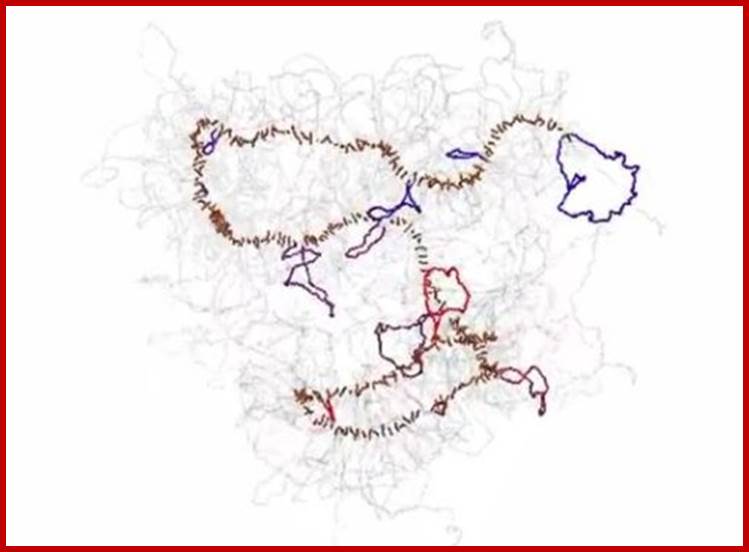
Prof.Job Dekker; http://www.sciencedaily.com/
This new insight into how chromosomes are disassembled and reassembled during cell division will allow researchers to begin answering basic questions about epigenetic inheritance, as well as human disease such as chromosome disorders and cancer; during metaphase the chromosome was being packaged in a two phase process. In the first phase, chromatin loops of 80,000 to 120,000 DNA base pairs form, radiating out from a scaffold and compacting the chromosome linearly. This was followed by axial compression of the chromosome, much like a spring being compressed, resulting in a neat, tightly folded package. DNA molecules are coiled up hierarchically into successively thicker fibers to ultimately form the sausage-like mitotic chromosomes; DNA forms a series of loops that are then attached to a linear axial structure that forms the backbone of the chromosome, both are supposedly correct; Prof.Job Dekkerhttp://www.sciencedaily.com/

Structurin of chromosomes through topological condensing links; Condensin structures chromosomal DNA through topological links; (a) Condensin complexes may structurally reinforce chromosome arms into rigid bodies that can be moved by mitotic spindle microtubules connected to a single kinetochore. Loss of rigidity triggered by release of condensin would cause chromosome arms to get stretched and lag behind centromeres during segregation. (b) Condensin rings may link different chromosome segments by encircling one chromatid segment while binding directly to a second segment of the same chromatid. The topologically bound segment may be free to slide through the ring. (c) Alternatively, condensin rings may encircle both segments within their ring structure.; Sura Cuylen et al ;www.nature .com
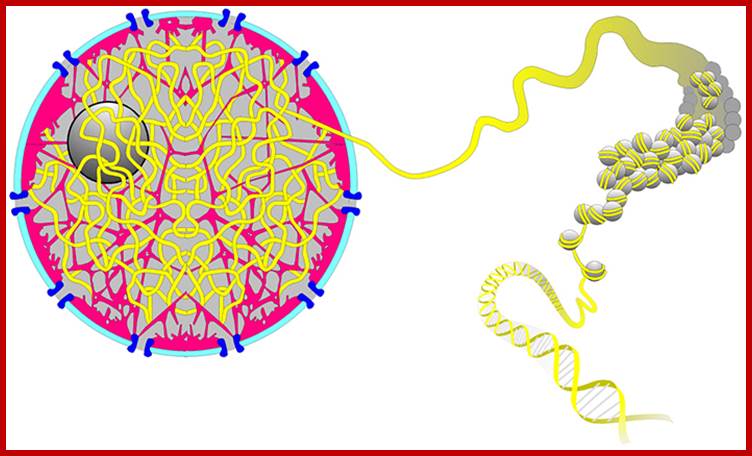
Levels of DNA organization in the nucleus. The DNA is wrapped around histone octamers to form the 10 nm chromatin fiber. This is condensed into the 30 nm fiber, which is in turn condensed into higher-order levels of organization. This highly condensed fiber is then packaged in the nucleus in an unknown manner. The nuclear matrix is represented in red. ;http://www.biology.emory.edu/
Gene expression and the three-dimensional organization of the genome:
Patterns of transcription required for cell differentiation during development are initially established by specific transcription factors. The maintenance of these patterns of gene expression is then carried out by alterations in chromatin structure that are epigenetically inherited between cell generations. These changes in chromatin organization take place at the level of the 10 nm fibre and include covalent histone modifications, alteration of nucleosome positioning by ATP-dependent remodelling complexes, and DNA methylation. In addition, more recent evidence suggests that the three-dimensional organization of the genome within the nucleus of eukaryotic cells may also be critical for achieving proper spatio-temporal patterns of gene expression during development. The factors and processes involved in the establishment, maintenance and regulation of specific states of nuclear organization are largely unknown but insulators are emerging as likely candidates to play this crucial role.
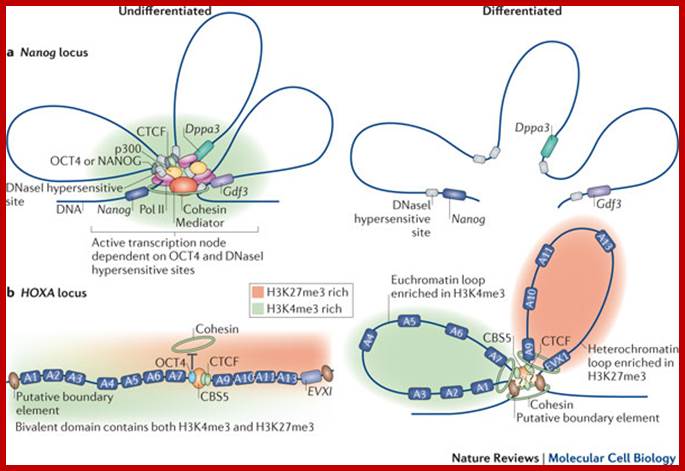
Models Depicting the higher-order chromatin structure in the nanolog ; OCT4 and CTCF organize three-dimensional chromatin loops in ES cells. Mo Li, Guang-Hui Liu & Juan Carlos Izpisua Belmonte; http://www.nature.com/

Transcription hub model for promoter choice of members of the Pcdhaa gene cluster in SDK-N-SH cells.;CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice; http://www.pnas.org/
Loops of chromatin; The first chromatin found to have an unusual looping 3D structure is a particular group of five genes making the beta subunit of hemoglobin. Genes sit on a loop right near the large regulator molecules needed to start and stop their production (promoters, enhancers and repressors). Loops can be flexible and the contact of the sites can be intermittent. This loop region makes it much easier to use the DNA. Often these loops create the environment for the activity, but a further stimulus is, also, needed.
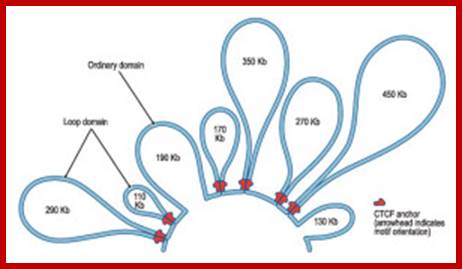
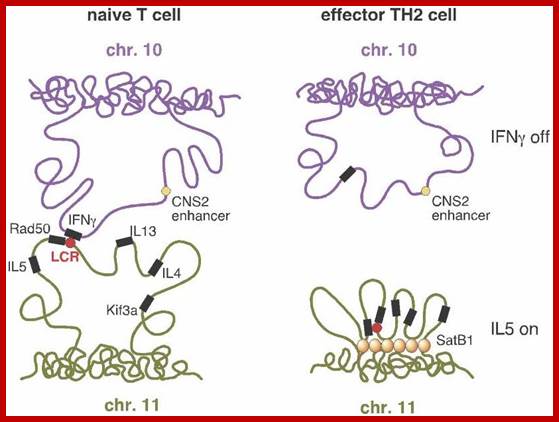
From Schneider and Grosschedl 2007.;http://natureinstitute.org/
The illustration (right side) shows the case
where a TH2 cell has resulted. Interferon (IFN-gamma) is not expressed![]() (upper right) because the looping pattern
separates the requisite gene from its enhancer
(upper right) because the looping pattern
separates the requisite gene from its enhancer![]() . On the other hand, IL-5 is expressed (lower right), because the
protein SATB1, which plays a large role in chromatin
. On the other hand, IL-5 is expressed (lower right), because the
protein SATB1, which plays a large role in chromatin![]() organization and transcription
organization and transcription![]() regulation, has anchored a series of chromosome
loops in just such a way as to bring the IL-5 gene and Rad50 promoter
regulation, has anchored a series of chromosome
loops in just such a way as to bring the IL-5 gene and Rad50 promoter![]() into proximity with the locus control region.
(Spilianakis et al. 2005; see also commentary in Kioussis 2005.)
into proximity with the locus control region.
(Spilianakis et al. 2005; see also commentary in Kioussis 2005.)
Silencing chromatin: comparing modes and mechanisms:
Recent transcriptome analyses show that substantial proportions of eukaryotic genomes can be copied into RNAs, many of which do not encode protein sequences. However, cells have developed mechanisms to control and counteract the high transcriptional activity of RNA polymerases in order to achieve cell-specific gene activity or to prevent the expression of deleterious sequences. Here we compare how two silencing modes — the Polycomb system and heterochromatin — are targeted, established and maintained at different chromosomal locations and how DNA-binding proteins and non-coding RNAs connect these epigenetically stable and heritable structures to the sequence information of the DNA.
Mode for the effect of SUMO modification on chromatin domain formation. Role of GYPSY insulators establish loop domains and insulators stabilize them Sumoylation of CP190 and Mod (mdg4)2.2 interferes with their self interactions resulting in the breakdown of chromatin domains. Gray line indicates nuclear lamina; http://emboj.embopress.org/
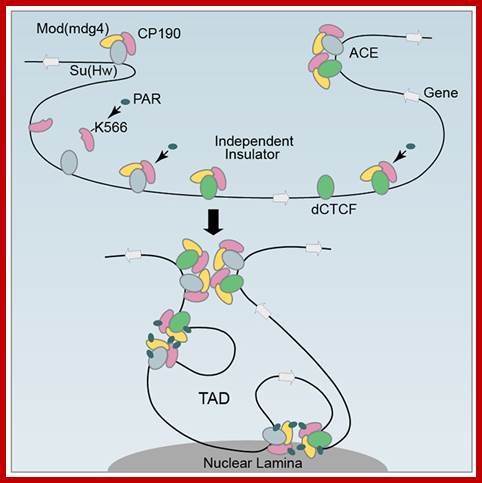
Model of how PARylation regulates insulator-mediated chromosome organization. PARylation of K566 in CP190 promotes its interaction with dCTCF. PARylation modulates the binding of insulator proteins within TADs, which in turn affect the intra-chromosomal interactions between distant insulator sites and their association with the nuclear lamina. Insulators play a role in the global 3D organization of the Genome: http://www.biology.emory.edu/research/
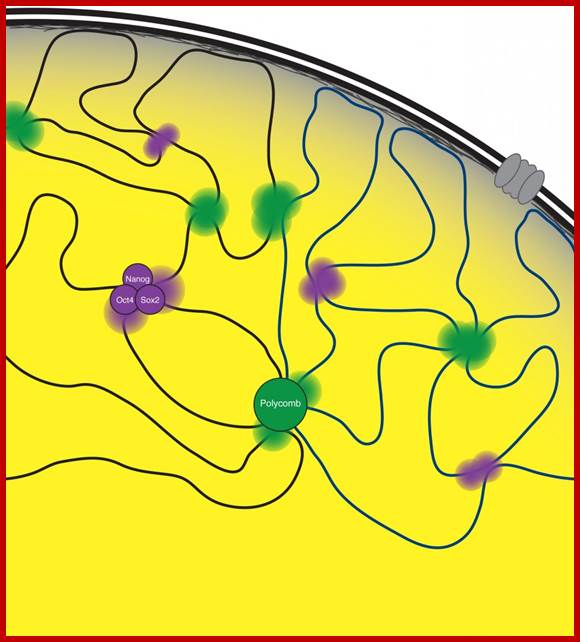
Regulation of gene expression during development and reprogramming; The DNA of eukaryotic cells is organized in a non-random fashion that allows for the accessibility of the gene regulatory machinery and for transcriptional and epigenetic changes that accompany cell fate changes. We discovered that transcription factors and chromatin regulators play a key role in bringing together stretches of DNA from distant sites in the genome, providing a mechanism for how co-regulated regions co-localize in 3D in the nucleus (Denholtz et al 2013). We are now developing models that comprehensively describe this spatial organization of the genome and the underlying molecular mechanisms; explore how this spatial organization of the genome contributes to the control of gene expression, and how changes in genome organization control cell fate transitions, cellular function, and disease processes. A second intriguing aspect of genome organization was revealed by our recent work on the mechanism of X-inactivation described above, providing an intricate link between genome organization and lncRNAs. For us, the X-inactivation paradigm therefore provides a unique opportunity to gain insights into the molecular mechanisms of genome organization, its dynamic regulation and relevance for gene and chromatin regulation.
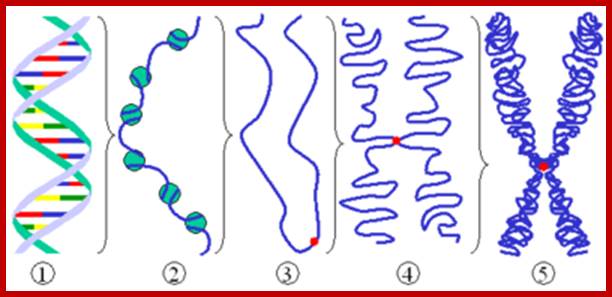
DNA goes through condensation into viable and visible chromosomes, www.wikibooks
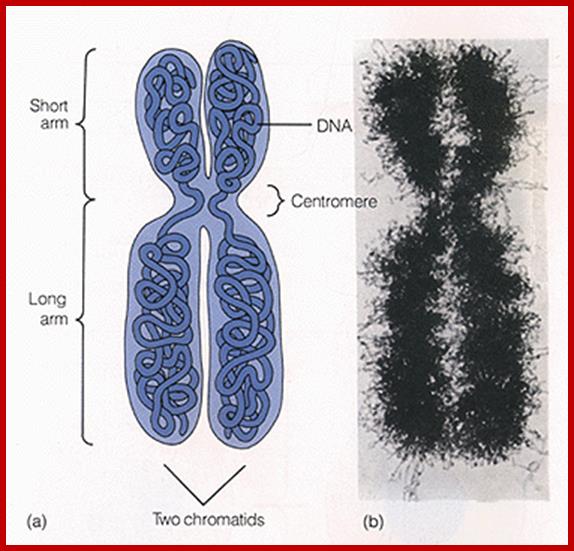
Unit 3 The Cell; https://sites.google.com
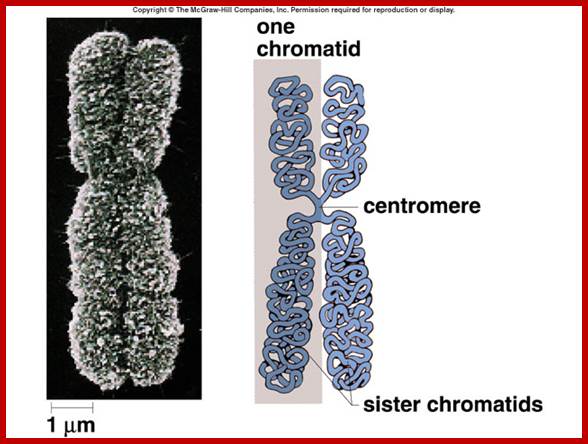
Organization and Control of Eukaryotic Genomes; https://gene-tics.wikispaces.com and Unit 3 The Cell ;https://sites.google.com
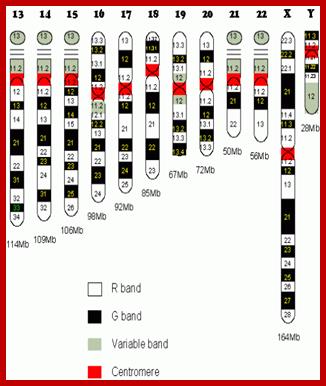

Human chromosomes and their genomic size; also shows structural bands; microbiology-sh.blogfa.com
Shape of the Chromosomes: Almost all chromosomes are spirally coiled thread like structures, but during cell division particularly at anaphase stage, chromosomes show a specific bent shape. This is due to the position of primary constriction or centromere. Accordingly, the chromosomes are called metacentric (V-shape) submetacentri (J-shape) acrocentric (rod shape),
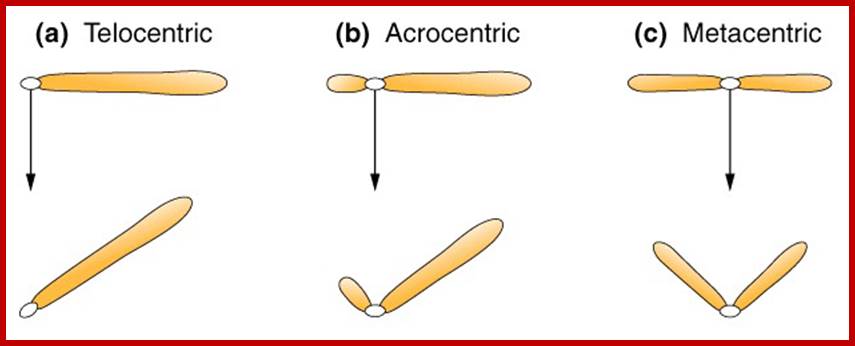
Telocentric (rod shape), polycentric (wave shape) and diffused (rod shape but move horizontally).www,mun.ca
Sex chromosomes and Autosomes: Higher organisms like man, monkeys and some plants where male and female sexes are morphologically differentiated, the cells in them contain two types of chromosomes, called Autosomes and sex chromosomes. The latter classification is based on the X and Y chromosomes. This classification is based on the chromatin nature and function. For example: human beings (Homo sapiens) have 46 chromosomes of which 44 are autosomes and 2 are sex chromosomes. Autosomes are believed to control the development of somatic body and sex chromosomes are responsible for the expression of sexual characters. If two XX chromosomes are present, female character is expressed; if one X and one Y chromosomes are present, male character is expressed. The X chromosome is considered to express female character and Y and the male character. The X chromosomes are euchromatic and Y are heterochromatic. Furthermore, the sex expression varies in different organisms. All in all, it is the interaction between autosomes and sex chromosomes that ultimately determines the expression of sexes through the mediation of specific hormones like Eaestrpgens (Female hormones) and Androgens male hormones).
Chromosomal structure
Using optical microscopes, if chromosomes of metaphase (at which chromosomes are at maximum condensation) are observed, chromosomes appear to be double stranded threads which are relatively coiled to each other. These threads are referred to as chromatids or chromonemata. If such chromatids are carefully observed under high resolution light microscope (2000 times enlarged), each of them appears to be spirally coiled with apparent gyrations. Many chromosomes show differential condensation, because of which some parts take more stains and other less stain. The former is called heterochromatic segments and the later are euchromatic. This differential staining behavior is called heteropycnosis. The heterochromatic segments may be either tightly coiled (take more strain) or less coiled (takes less stain), this feature is called as positive heteropycnosis and negative heteropycnosis respectively.
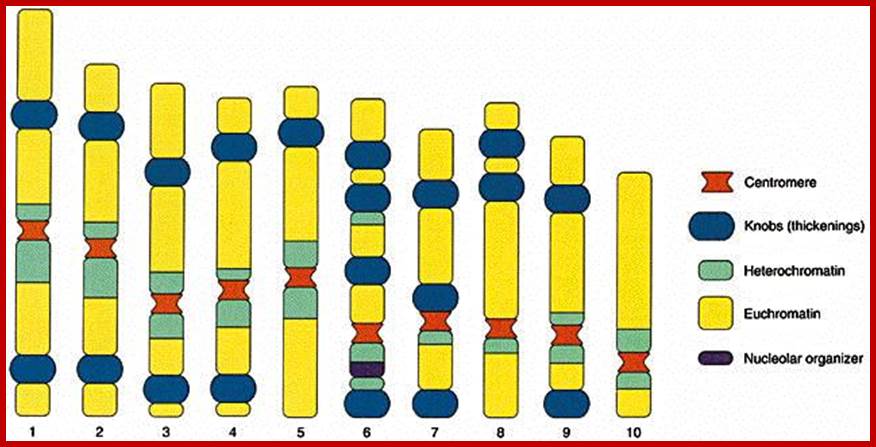
In some cases the entire chromosome appears to be heterochromatic because of greater condensation. The heterochromatin can be constitutive in CEN and telomeric regions and it can be facultative also, the position of such constitutive regions change during development and from one tissue to the other. Formerly, these heterochromatic regions were believed to be genetically inert, now they are known to contain genes and they do express. Furthermore, the heterochromatic region is not constant and same in all the cells of an organism. However, now, it is known for certain that some heterochromatic region contains redundant DNA segments.
The inheritance of Mendelian factors or characters through chromosomes was not substantiated till the discovery of chromosomes. This has, however, led to unit character inheritance, specially located within chromosomes. These unit characters are now called genes. Nevertheless, the problem of genes which are arranged in chromosomes was an enigma, but the discovery of bead like structures in meiotic chromosomes, called chromomeres, has given an impetus in unraveling this problem. Meiotic chromosomes particularly at leptotene stage, appears as fine strings of beads.
These bead like structures were called as chromomeres and were equated to individual genes. Such chromomeres were equated held by nongenetic threads. But later, chromomeres were found none other than the coiled expressions of chromonemata; when two ends of such chromatids are stretched apart, the chromomeres disappear. Nevertheless, the concept of linear arrangement of genes in chromosomes has been accepted. Morgan and later Hunt’s cytogenetic experiments have furthered the concept of gene as a unit of heredity, a unit of recombination, a unit of mutation and a unit of function.

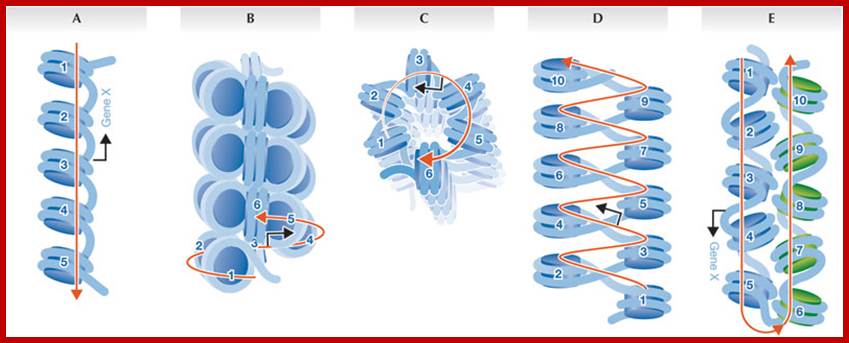
Models- A. 10nm fibre, B. Side view of30nm fiber, C. Top-Down view, D. Zig-Zag model of 30nm fiber, E. Inter digitation of two 10nm fibers; Number of circles are nucleosomes in an array; http://embor.embopress.org/
Morphologically the metaphasic chromosomes appear to be simple, spirally coiled threads of uniform thickness; here and there the chromosomes contain constricted or narrow regions. These are called primary constrictions and secondary constrictions respectively. The primary constrictions and secondary constrictions are further differentiated and characterized by their behavior and functions.
Centromere: The primary constriction is also called centromere for it is the region at which chromosomes get attached to tractile fibres and they lead the anaphasic chromosomal movements. If the centromere is destroyed by direct X-radiation hits, chromosomes behave abnormally and lose their directional movements. Thus centromere appears to be a non stainable gap, the chromosomal thread. However, in metaphasic chromosomes, though the arms are double stranded in the centromere region, the centromere still appears to be single stranded, but in actuality it is double stranded.
Centromere is attached to tractile fibers; www.dna20.com
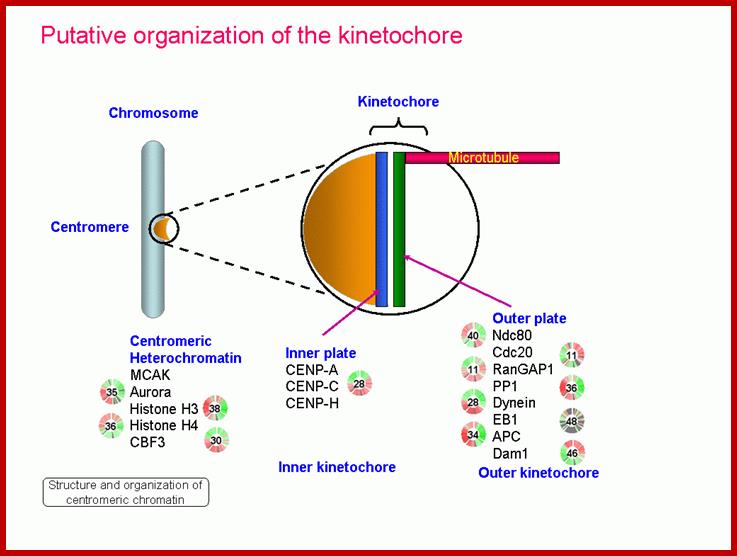
Kinetochore components; www.mpmp.huji.ac.il
Recent electron microscopic studies clearly show that either side of the centromeric region, in line with the chromosomal arms, is covered by or ‘C’ shaped structure called Kinetochore. Thus two kinetochore structures are present in each centromere. Each kinetochore is made up of three regions. The outer most region is cup shaped structure called commissural cup and it is relatively thick. On its outer surface large number of processes called corona are present. Amidst the corona a number of (3-10) microtubules are found to be penetrated as deep as to reach chromonema. The middle region is less dense, but the inner region is dense and it is in contact with centromeric chromonema. However, the entire kinetochore structure appears to be a centromeric gene product and it is likely to participate as the nucleating centre in the polymerization of tubulin into tractile fibres during the formation of mitotic apparatus.
The position of centromere for a given chromosome in a given set of chromosomes is specific and characteristic. It may be metacentric (middle), submetacentric (little away from middle), acrocentric (at the extreme tip but with a small segment at the end) and telocentric (extreme tip). In some cases two or more centromeres are found in the same chromosome (Dicentric or polycentric) but in certain cases the centromeric activity is found throughout the length of chromosome and it is called diffused centromere. The shape of the chromosome depends upon the position of the centromere.
Another interesting feature of the centromere is that, on either side of the centromere the chromatin material is heterochromatic. And it is believed that the genes in this region are highly redundant, however their function is not known.
Secondary constriction or Nucleolar organizer: Similar to that of primary constriction another constriction is present on only one or two pairs of specific chromosomes. Such constriction is called secondary constriction and it is characterized by its nucleolar formation, hence it is also referred to as Nucleolar organizer. This region contains DNA segments for cytoplasmic ribosomal RNA. The genes present, here, are redundant or tandem repeats. In some cases, like Frog oocytes these genes get amplified at the end of the telophase. The DNA in the region opens out, and starts transcribing ribosomal RNA as large precursors. Later this RNA is sliced and processed into 28S and 5.8S RNAs. These RNAs in turn get associated with different ribosomal proteins sequentially and functional ribosomes are produced. If the secondary constriction segment of the chromosome is knocked off, the nucleolus fails to appear and the cells die prematurely.
SAT Chromosomes; Short chromosomal segments at the terminal region beyond secondary constrictions are called Satellite. Such chromosomes are called SAT chromosomes. Heitz, who coined this term, is in the opinion that these segments are lacking in thymidilic acid, i.e. Sine Acido Thymidine (SAT). Otherwise, this region, as it is lacking Thymidilic acid, it is rich in Guanidilic and cytidilic acids. Whether such GC rich heterochromatic chromosomal segments contain some redundant genes or not, is not known. Their function is also a mystery.
Telomeres: In most of the plants, the chromosomal ends have heterochromatin materials. Such structures are referred to as telomeres. The presence of such structures are found to be non-sticky and prevent
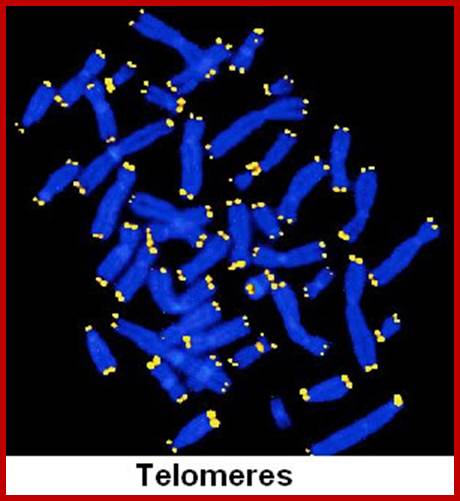
Telomeres are specifically labelled; www.planetatheism.com
the attachment of chromosomal ends. If telomeres are cut off, the ends become sticky. However, the recent in situ hybridization techniques have demonstrated that heterochromatic telomeres contain redundant DNA segments, coding for 5.8S r.RNA of large subunit of cytoplasmic ribosomes.
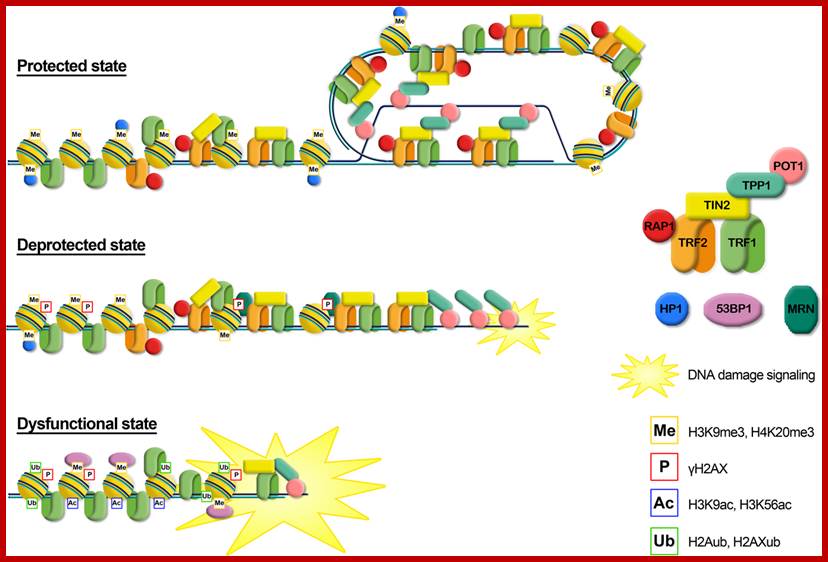
Graphical representation of the different telomere states, characterized by different levels of telomeric proteins and post-translational modifications. Protected state: telomere is in a closed form, probably the t-loop, maintained by the binding with the shelterin proteins; the presence of trimethylation of histones H3 and H4, typical heterochromatic markers, induces a compacted state. This state inhibits the DNA damage response. Deprotected state: telomere shortening could disrupt the closed structure leading to an open state, characterized by a decrease of heterochromatic marks. Telomeres are recognized as DNA damage, signaled by phosphorylation of H2AX, but retain enough shelterin proteins (mainly TRF2) to prevent NHEJ and thus telomeric fusion. DNA damage signaling leads to replicative senescence. Dysfunctional state: if growth arrest checkpoint is inactivated, telomeres continue to shorten leading to a fully uncapped form, deriving from the depletion of shelterin proteins such as TRF2 or POT1. Telomere dysfunctions are signaled by phosphorylation of H2AX and the ubiquitylation of H2A and H2AX. Telomeres are not protected from the DNA damage response machinery, giving rise to extensive telomere fusions. http://journal.frontiersin.org/
Ultrastructure of Chromosomes
Observations of metaphasic chromosomes under optical microscopes of reasonable resolution, shows them as fine, spirally coiled threads. Beyond this, it is difficult to understand the internal organization of chromosomes. Nevertheless the techniques of whole mount chromosomes combined with electron microscopy, have revealed that chromosomes are made up of highly folded but spirally coiled chromonemal threads of 250-300 A0 thickness, with ends visible nowhere.
Meanwhile, biochemical studies have revealed that the chromosomes contain DNA, histone and non-histone proteins. The complexity of the organization of the above said components has further compounded with the discovery of various components of histones and nonhistones. Histones are found to be basic proteins and different types have been identified, viz., H1, H2A, H2B, H3, and H4.
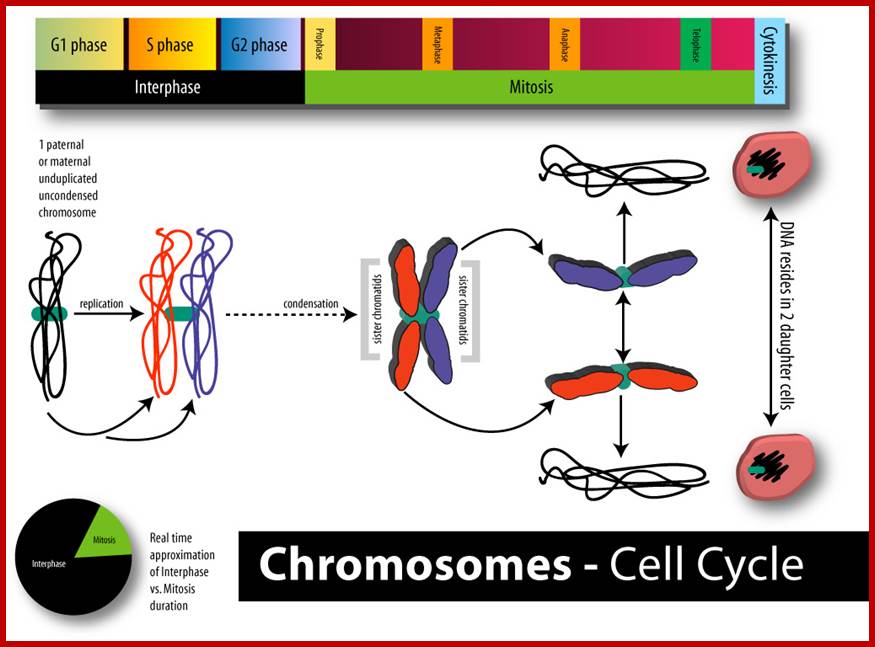
Notwithstanding this, nonhistone proteins are found to be acidic in nature and 100-120 or more different kinds have been suspected to be present. At the top of it, a single chromonema has been found to contain a single DNA molecule.
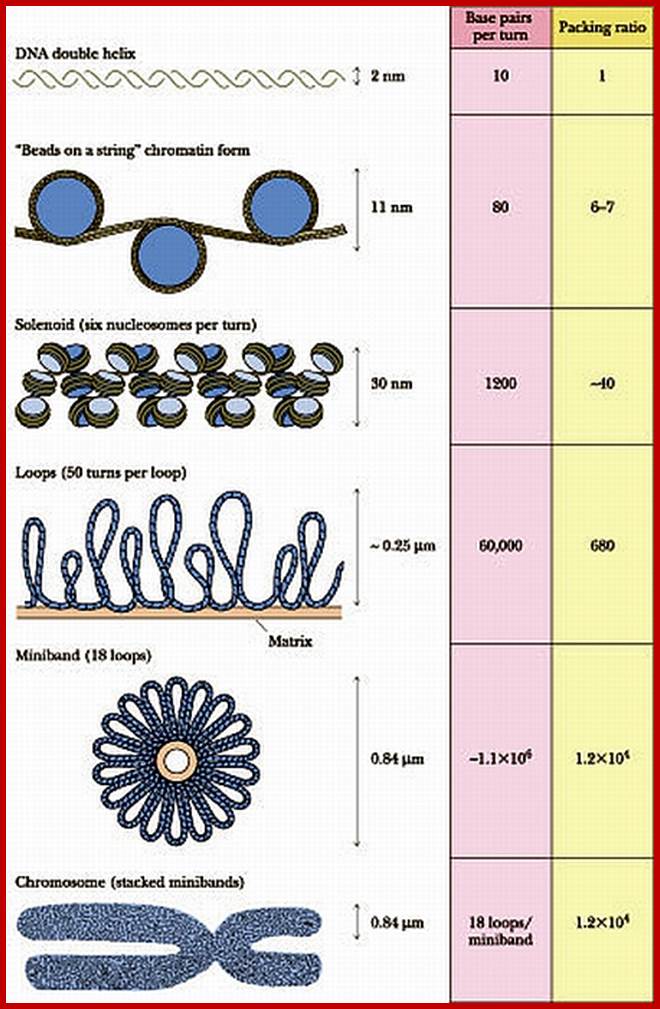
www.wmin.org;www.Nature.com
When chromatin is added to a salt medium, the chromatin threads spread into fine bead like structures called Nu-bodies or Nucleosomes. The Nu-bodies were further identified as to contain histones as octamer (2H2A 2H2B, 2H3 and 2H4) and DNA double helix of about 140 base pairs length is coiled around it. The DNA thread found in between two such Nu-bodies is called linker DNA which consists of 50 to 100 base pairs. Furthermore, the electron microscopic studies of polytene chromosomes reveal that the Nu-bodies or nucleosomes are biconvex disc shaped structures with histone octamer as the core, around which DNA coiled 1 ¾ times or twice. These structures have been recognized as the fundamental units of chromosomal fibers.
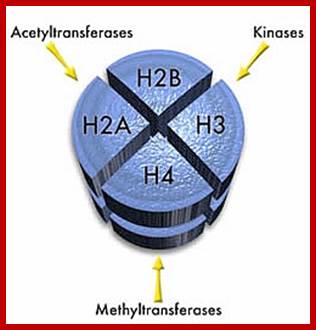

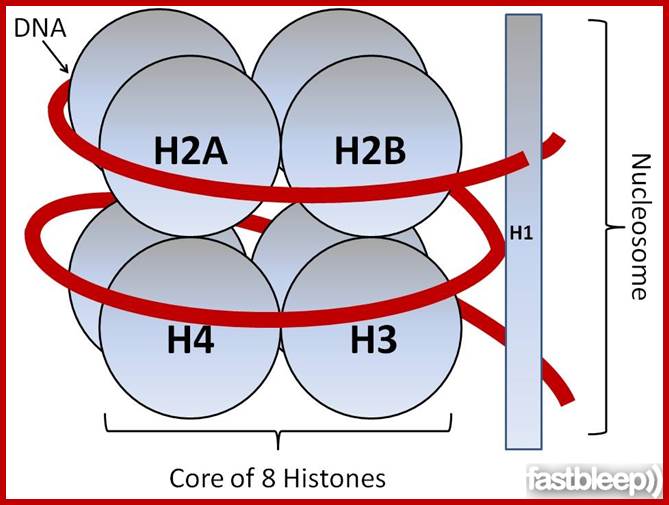
http://www.fastbleep.com
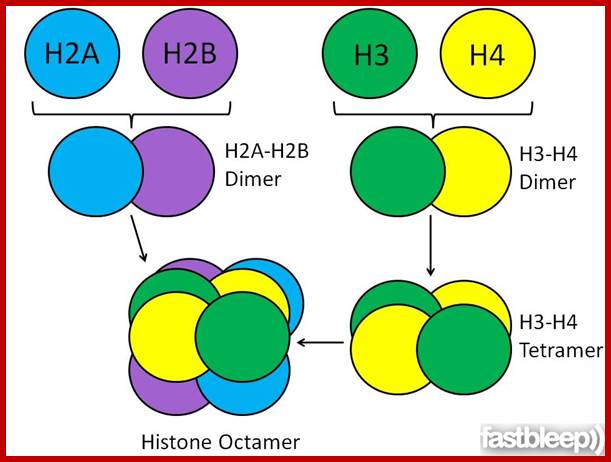
http://www.fastbleep.com/http://themedicalbiochemistrypage.org/
Such fundamental chromosomal fibres with Nu-bodies as units, coil round each other with H1 proteins and others as the binding forces to a thread of first order called Solenoid structure. Then this solenoid thread undergoes further coiling to produce a super coiled thread of 400 nm thickness, where the wall of the coil retains the same 70 nm thickness. These are called chromonema which is visible under light microscope. These threads show a contraction of 1300 to 1500 fold to that of string of beads. Such chromonemata are further coiled in metaphasic chromosome which exhibits a 6 x 103 – 7 x 103 fold condensation. However the coiling and condensation of Nu-bead fibres into microscopically visible chromosome, is aided and augmented by a number of non-histone proteins as binding factors to form a kind of scaffold. If such chromosomes are subjected to histone digestion and dispersed under certain detergent cum salt solution the histones are selectively removed, but retain the non-histone proteins intact. All the histone free DNA molecules spill out in the form of fiber loops.
Artificial or synthetic chromosomes: Arrow; banats-quotes-pickups.blogspot.com
First synthetic Yeast Chromosome for complex-cell organism: Researchers took tiny snippets of man-made DNA and joined them together to create a synthetic version of a chromosome, the structure that contains DNA inside cells, from brewer’s yeast. The ability to create such chromosomes is a major step for the field of synthetic biology, which aims to engineer microbes to produce useful products. The work also brings scientists closer to creating synthetic plants and animals http://www.sciencebeing.com/
These and other studies, notwithstanding the complexity of the association of DNA with proteins, have shown that each chromonemata is made up of single, but a long DNA molecule, which supports the concept that the genes are arranged in linear order, where a segment of DNA acts as the unit of heredity.
Giant chromosomes
Chromosomes, in most of the organisms show a cycle of condensation and decondensation during various stages of cell divisions. But in some organisms, enlarge considerably to the tune of 250-500 times the normal somatic chromosomes. These are called giant chromosomes and they are not just restricted to one species but found to occur in various organisms like insects, frogs, salamanders and even plants. However, their occurrence is restricted to certain stages in the life cycle. Two such giant chromosomal types’ i.e. salivary gland chromosome and lampbrush chromosome have been extensively studied.
Salivary gland chromosomes or polytene chromosomes: Salivary gland chromosomes are found in salivary gland cells of 11th day larvae of dipteron class of insects. They are present in the larvae of Chironema, Drosophila melanogaster, mosquitoes and other insects. Such giant chromosomes are also found in the endosperm haustorial cells of Phaseolus vulgaris.
In the body of these insects, these chromosomes remain normal in their size, but when the larva reaches the 11th day and is about to undergo metamorphosis into pupa, the somatic chromosomes found in the salivary glands undergo a dramatic change in the size and activity. At this stage of development, chromosomes undergo repeated chromosomal divisions without separation, which leads to the multiplication of two chromatin threads into 1000-1024 strands. Furthermore, these somatic homologous pairs of chromosomes are found to be in pairs or synapsis.
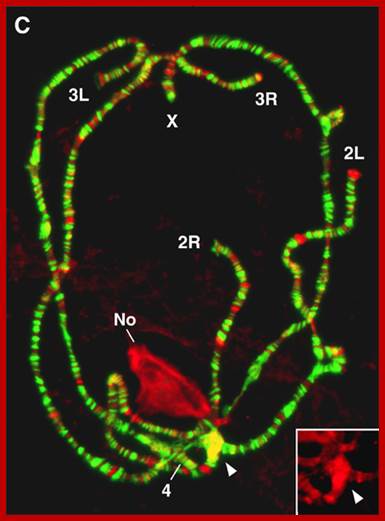
Salivary gland chromosomes of Drosophila
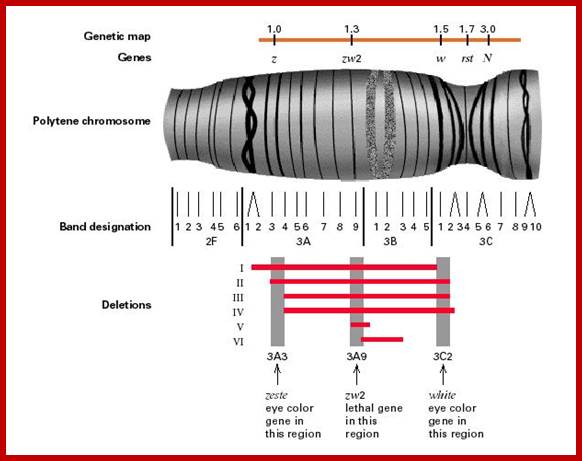
Structurally these chromosomes contain thousands of genes longitudinally oriented chromonemal threads arranged parallel to each other. These threads also show cross banding of various sizes. Some bands take greater stain and others stain less. The Darkly stainable bands are heterochromatin segments where chromosomes are densely packed with chromatin material.
Occasionally some of these darker bands containing highly condensed chromatin uncoil and open out and start transcribing specific m.RNAs. The transcriptional activity has been identified by the use of radioactive precursors. This region represents intense gene activity. Because of this reason, this region appears be puff like structure which consists of opened out DNA strands in the form of a ring like structures called Balbiani rings. Such puffs can be induced in the chromosomes by applying an hormone called ecdysone. This hormone induces the transformation of larva into pupa, during which many morphological structures and functional activities of the larva undergo dramatic changes. Probably, because of these reasons, the 11th day larval chromosome undergoes differential gene activation to synthesize required protein products for the metamorphosis.
Lampbrush chromosomes: Chromosomes which appear as bottle brush or the brush that is used to clean the lamp glasses are called lampbrush chromosomes. These chromosomes are highly elongated (5900 um) and they are found in the oocytes, the homologous pairs of chromosomes undergo enlargement and differentiation. To begin with, the homologous pars undergo synapsis, and later they are held to each other at charismatic regions.
Lampbrush chromosomes; http://www.cbs.dtu.dk/
Each homologous chromosome consists of two chromonemata containing a large number of granular structures called chromomeres. These chromomeres, all along the length of chromatin threads during diplotene open out in the form of large lateral loops. Soon, these loops will be covered by a matrix, which is made up of a pool of RNA nucleotides and RNA polymerases. These lateral loops now start intense transcriptional activity, with the result, innumerable RNAs are synthesized. Each loop consists of many genes of the same kind separated by noncoding regions called spacer DNA. Transcriptional activity is initiated at several sites simultaneously by the binding of RNA polymerases and soon RNA strands are formed with the progression of RNA polymerase. As soon as the initiating site is free, another RNA polymerase settles and starts synthesis at various stages and each of these segments appears as the branch of a christmas tree. RNA synthesis is required for the formation of ribosomal structures. Some of the RNAs thus produced are rRNAs. Thus a large number of ribosomes and m.RNAs are stored in the eggs. This is another instance of differential gene activity, required for certain development process.
Disassembly and reassembly of Nuclear membrane during cell division:
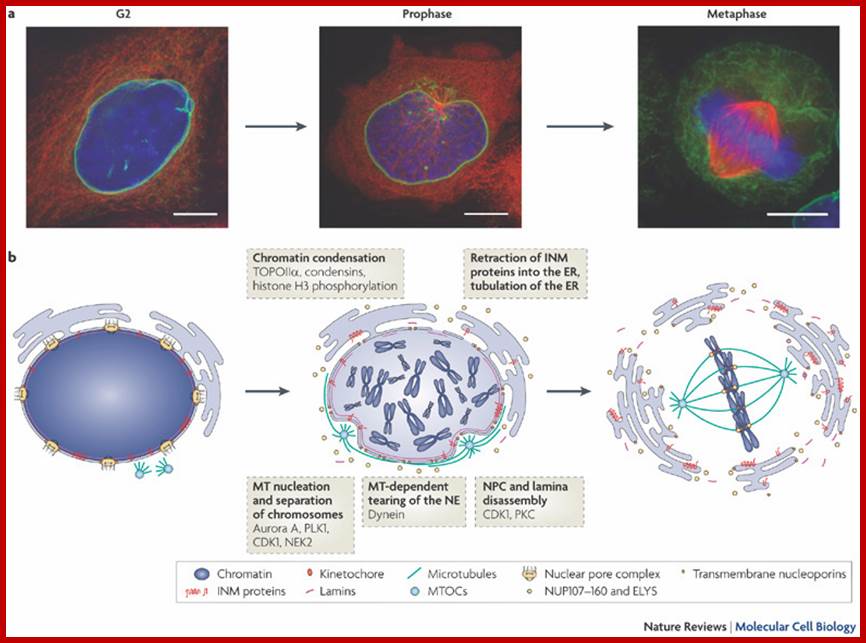
Nuclear envelope breakdown during 'open' mitosis. And Orchestrating nuclear envelope disassembly and reassembly during mitosis Stephan Güttinger, Eva Laurell & Ulrike Kutay; Nature Reviews Molecular Cell Biology-
a | The
images show HeLa cells in which the inner nuclear membrane (INM, green; stained
by green fluorescent protein fused to lamina-associated protein 2![]() ), DNA (blue; stained with Hoechst) and
microtubules (red; stained by red fluorescent protein–
), DNA (blue; stained with Hoechst) and
microtubules (red; stained by red fluorescent protein–![]() -tubulin) are visualized in G2, prophase and
metaphase. Scale bars, 10
-tubulin) are visualized in G2, prophase and
metaphase. Scale bars, 10 ![]() m. b | At the
end of G2 phase, the activation of mitotic kinases, including the master
mitotic regulator cyclin-dependent kinase 1 (CDK1), triggers entry into
prophase, which is associated with a series of events that include the start of
chromatin condensation, formation of microtubule asters around centrosomes and
centrosome separation. Microtubules that are attached to the nuclear envelope
(NE) in conjunction with the minus-end-directed motor dynein lead to NE
invaginations around centrosomes and to the formation of holes on the opposing
site of the NE. At the same time, nuclear pore complex (NPC) disassembly
commences and is probably caused by the phosphorylation of nucleoporins. The
transition into prometaphase is marked by the loss of the NE permeability
barrier. Phosphorylation of nuclear lamins and INM proteins by CDK1, protein
kinase C (PKC) and probably other kinases results in lamina disassembly and
allows for the retraction of NE membranes into the endoplasmic reticulum (ER).
In metaphase, most soluble components of the NE are dispersed throughout the
cytoplasm, whereas INM proteins reside in the tubular mitotic ER. ELYS is known
as MEL-28 in Caenorhabditis
elegans. MTOC, microtubule-organizing centre; NEK2, NimA-related
kinase 2; PLK1, polo-like kinase 1; TOPOII
m. b | At the
end of G2 phase, the activation of mitotic kinases, including the master
mitotic regulator cyclin-dependent kinase 1 (CDK1), triggers entry into
prophase, which is associated with a series of events that include the start of
chromatin condensation, formation of microtubule asters around centrosomes and
centrosome separation. Microtubules that are attached to the nuclear envelope
(NE) in conjunction with the minus-end-directed motor dynein lead to NE
invaginations around centrosomes and to the formation of holes on the opposing
site of the NE. At the same time, nuclear pore complex (NPC) disassembly
commences and is probably caused by the phosphorylation of nucleoporins. The
transition into prometaphase is marked by the loss of the NE permeability
barrier. Phosphorylation of nuclear lamins and INM proteins by CDK1, protein
kinase C (PKC) and probably other kinases results in lamina disassembly and
allows for the retraction of NE membranes into the endoplasmic reticulum (ER).
In metaphase, most soluble components of the NE are dispersed throughout the
cytoplasm, whereas INM proteins reside in the tubular mitotic ER. ELYS is known
as MEL-28 in Caenorhabditis
elegans. MTOC, microtubule-organizing centre; NEK2, NimA-related
kinase 2; PLK1, polo-like kinase 1; TOPOII![]() , topoisomerase II
, topoisomerase II![]() . Eva Laurell & Ulrike Kutay
. Eva Laurell & Ulrike Kutay
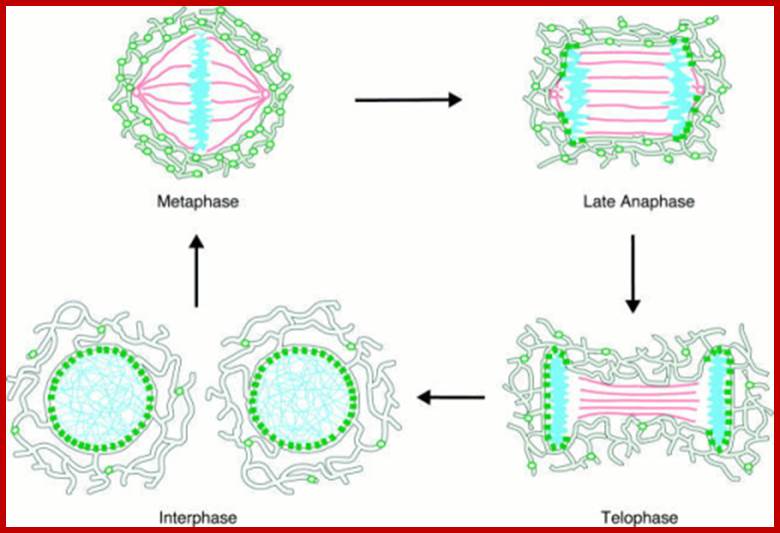
Nuclear Membrane Dynamics and Reassembly in Living Cells: Targeting of an Inner Nuclear Membrane Protein in Interphase and Mitosis; Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J - J. Cell Biol. (1997)
Modelof Nuclearenvelopereassembly.
Model of nuclear envelope reassembly. In interphase (bottom left), newly synthesized integral inner nuclear membrane proteins such as LBR (green ovals) move by lateral diffusion from the ER (gray outlined network) to the inner nuclear membrane, where they are retained and immobilized (green squares) by binding to nucleoplasmic ligands (chromatin, blue). Early in mitosis, these binding interactions are disrupted, leading to equilibration of the mobilized LBR molecules within the ER/NE system. In metaphase (top left) the NE is completely disassembled and LBR diffuses freely within an interconnected ER network that surrounds the spindle apparatus (red) and the condensed chromatin. Binding sites for LBR are available again in late anaphase (top right), immobilizing the receptor at contact sites between ER and . Cell Biol. (1997)
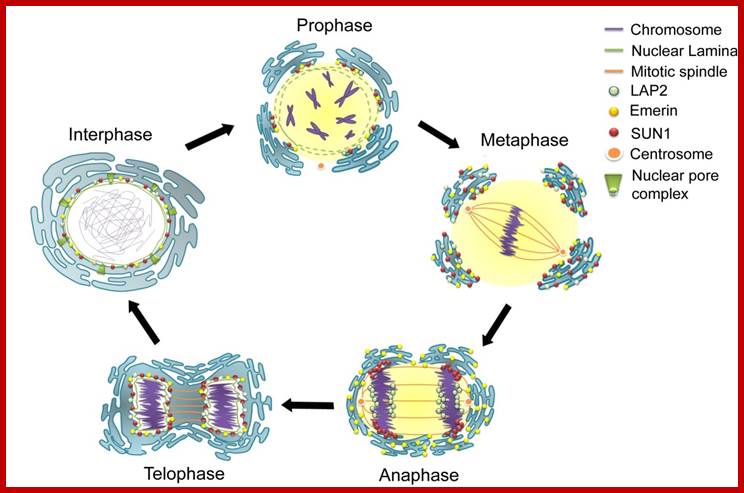
Nuclear envelope breakdown and reassembly in mitosis. At the end of G2, the activation of cyclin-dependent kinases, including CDK1, triggers entry into mitotic prophase. The nuclear membrane breaks down, and the NE-associate proteins either translocate to kinetochores, distribute with the fragmented ER networks, or dissolve in the cytoplasm. During NE reassembly in anaphase, SUN1 and LAP2 first appear around the condensed chromatin, though at different regions. The nuclear lamins then join the nuclear periphery in telophase. This figure illustrates the important roles played by the NE and the nuclear lamina in normal mitosis. Chi et al. Journal of Biomedical Science 2009
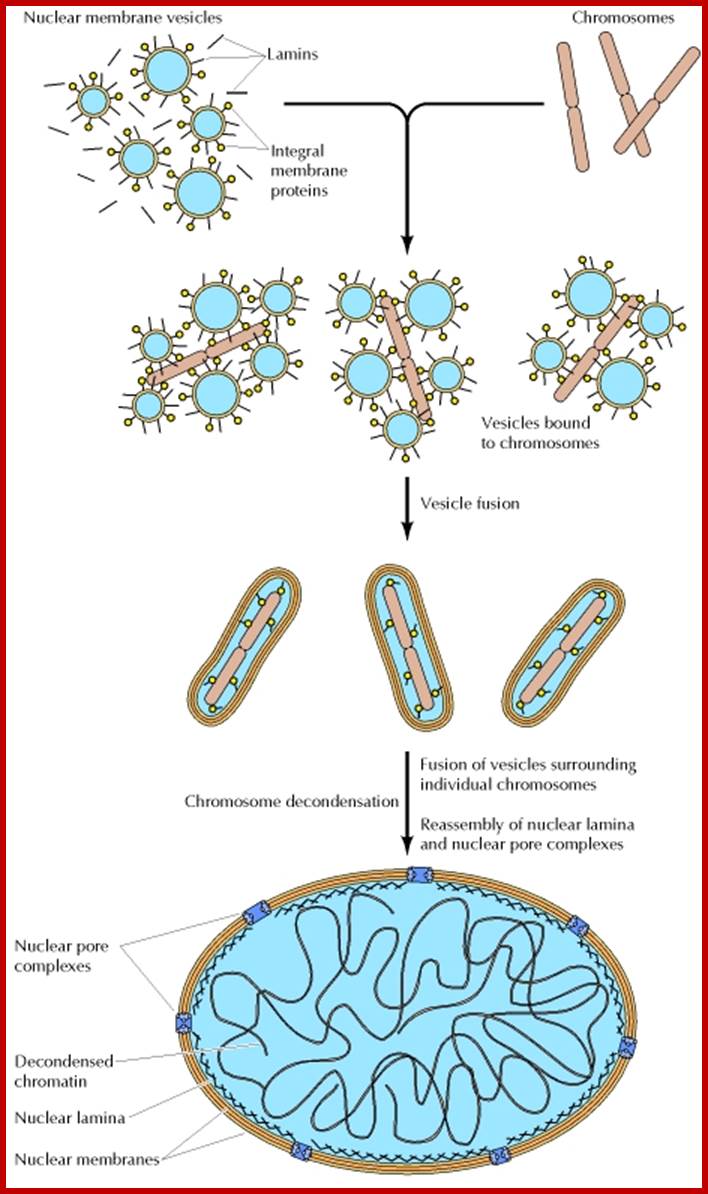
Re-formation of the nuclear envelope. The first step in reassembly of the nuclear envelope is the binding of membrane vesicles to chromosomes, which may be mediated by both integral membrane proteins and B-type lamins. The vesicles then fuse, the nuclear membrane reforms. The first step in reassembly of the nuclear envelope is the binding of membrane vesicles to chromosomes, which may be mediated by both integral membrane proteins and B-type lamins. The vesicles then fuse, the nuclear lamina reassembles, and the chromosomes decondense. http://www.ncbi.nlm.nih.gov/
The mechanisms of localization and retention of membrane proteins in the inner nuclear membrane and the fate of this membrane system during mitosis were studied in living cells using the inner nuclear membrane protein, lamin B receptor, fused to green fluorescent protein (LBR-GFP). Photo bleaching techniques revealed the majority of LBR-GFP to be completely immobilized in the nuclear envelope (NE) of interphase cells, suggesting a tight binding to heterochromatin and/or lamins. A subpopulation of LBR-GFP within ER membranes, by contrast, was entirely mobile and diffused rapidly and freely (D = 0. 41 +/- 0.1 microm2/s). High resolution confocal time-lapse imaging in mitotic cells revealed LBR-GFP redistributing into the interconnected ER membrane system in prometaphase, exhibiting the same high mobility and diffusion constant as observed in interphase ER membranes. LBR-GFP rapidly diffused across the cell within the membrane network defined by the ER, suggesting the integrity of the ER was maintained in mitosis, with little or no fragmentation and vesiculation. At the end of mitosis, nuclear membrane reformation coincided with immobilization of LBR-GFP in ER elements at contact sites with chromatin. LBR-GFP-containing ER membranes then wrapped around chromatin over the course of 2-3 min, quickly and efficiently compartmentalizing nuclear material. Expansion of the NE followed over the course of 30-80 min. Thus, selective changes in lateral mobility of LBR-GFP within the ER/NE membrane system form the basis for its localization to the inner nuclear membrane during interphase. Such changes, rather than vesiculation mechanisms, also underlie the redistribution of this molecule during NE disassembly and reformation in mitosis. http://openi.nlm.nih.gov/; Ellenberg J, etal.
