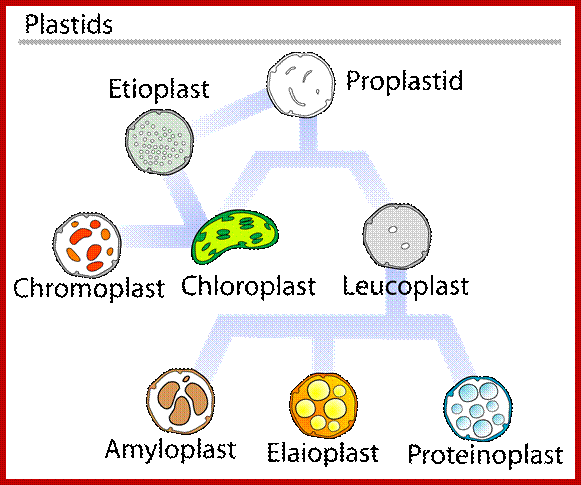
Plastids
Plastid is another important energy transducing cell organelle found only in plants. Shimper coined the name Plastids for those structures responsible for photosynthesis. In actuality, photosynthesis provides chemical energy directly or indirectly, for all other living organisms, Chloroplasts are unique organells for they are capable of capturing, converting and conserving solar energy in the form of chemical energy. Plastids are found in almost all cells of the plant body either in the form of colorless plastids or colored plastids or proplastids.
Classification:
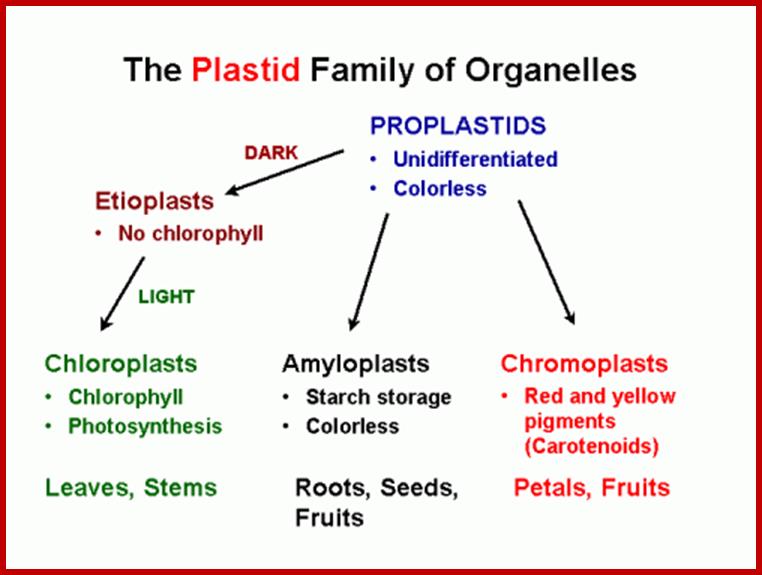
On the basis of presence or absence of pigments, and the stage of development, plastids have been classified into proplastids, leucoplasts and chromoplasts.
Proplastids:
Small vesicular structures present in meristematic cells are called proplastids. They are colorless and undeveloped. As cells mature into different cell types, depending upon the organs and presence or absence of light, proplastids undergo transformation and develop into either colorless leucoplasts or colored chromoplasts including green chloroplasts. Proplastids continuously divide and redivide and provide them for cells undergoing differentiation into various types.
Leucoplasts:
Colorless plastids that are found in storage parenchyma and other colorless tissues are refereed to as leucoplasts. Most of them act as storage organelles. Based on the kind of substance they store they are further classified into amyloplasts. If such leucoplasts are exposed to sunlight they will be transformed into colored plastids, which suggest that these plastids have retained all the genetic potentiality to develop and perform photosynthesis.
Chromoplasts:
All plastids containing different colored pigments are grouped under chromoplasts, of which green colored ones are called chloroplasts. Depending upon the dominant pigments present in plastids, they are further classified into Rhodoplasts rich in red pigment i.e. phycoerythrin. Phaeoplasts and Xanthoplasts contain yellow pigments i.e. xanthophylls, carotinoids. Along with the above pigments phycocyanin and other pigments are also present in other colored plastids.
Other plastids: Such colored plastids, other than chloroplasts are predominantly found in certain class of plants and plant organs including floral parts. Though floral parts are derived from the same set of proplastids, produce different pigments in petals. The exact process differentiation is not known for different plants do produce different colored petals and it is genetically programmed.
Interconversion:
Proplastids divide and redivide in meristematic cells, and then they are distributed to cell derivatives on exposure to light, depending upon the structures in which they found and also depending upon the intra cellular factors they develop into colorless plastids or colored plastids. Leucoplasts on exposure to light develop into green plastids. Similarly, chloroplasts may become leucoplasts; but colored plastids as in petals are mostly terminally differentiated.
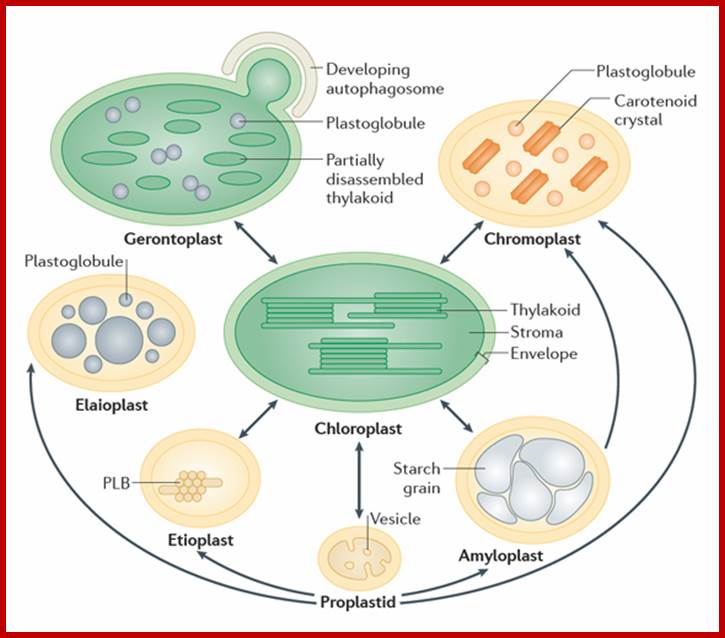
Plastids exist in different forms, and the identity and abundance of each are controlled by developmental and environmental cues. Different types interconvert (see the arrows) following reorganization of the organellar proteome, a process that is controlled by the differentially regulated import of nucleus-encoded proteins. Chloroplasts are photosynthetic plastids, Amyloplasts are starch-storing plastids, chromoplasts are carotenoid pigment-accumulating plastids, and proplastids are undifferentiated plastids that can differentiate into the different types of plastids. Etioplasts are chloroplast progenitors that form in darkness and accumulate chlorophyll precursors (in paracrystalline membranous structures called prolamellar bodies (PLB)) that are ready for rapid differentiation upon illumination. Elaioplasts store lipids in lipid droplets known as plastoglobules and exist, for example, in tapetal cells during pollen development. Gerontoplasts form during senescence, owing to resource recycling through the disassembly of the photosynthetic machinery and autophagy. Taken from Nat. Rev. Mol. Cell Biol., 2013, 14: 787-802.;http://users.ox.ac.uk/
www.imagercade.com
Chromatophores:
Prokarytic photosynthetic bacteria contain photosynthetic organelles called chromatophores. Bacterial chromatophores are made up on membranous vesicles in which photo synthetic pigments and other required factors are found. In these structure photosynthetic pigments, associated with light harvesting proteins and required enzymes are aggregated into photosynthetic units. Apart from that, chromatophores do now show any specialized structural differentiation. However, in blue green algae, the entire cell apart from its cell wall acts as a giant chloroplast with its grana and stromatic fluid.
Neill Lambert, et al; http://www.nature.com/
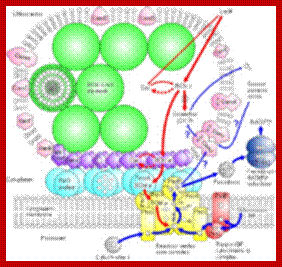
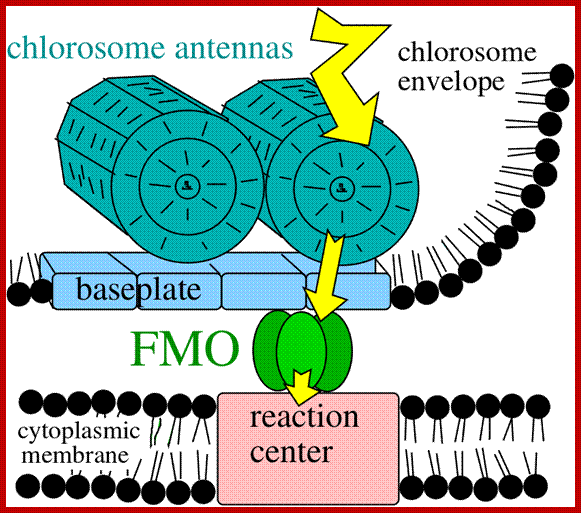
Model depicting the process of light harvesting in C. tepidum. The model includes the chlorosome, FMO protein, and reaction center. Photo courtesy of the University of Copenhagen Biology Department (http://www.bio.ku.dk/nuf/images/Cte
Quantum aggregates are assemblies of monomers (molecules, atoms, quantum dots...), where the monomers largely keep their individuality. Interactions between the monomers can lead to collective phenomena, like super radiance or efficient excitation transfer. Researchers study different kinds of aggregates (e.g., light harvesting systems, arrays of Rydberg atoms, self-assembled organic dyes). Using various methods, we study optical and excitation transport in these systems. Of particular interest is the coupling of the excitation to nuclear degrees of freedom.; Energy transfer in the photosynthetic Fenna-Matthews-Olson (FMO) complex of green sulfur bacteria is studied numerically taking all three subunits (monomers) of the FMO trimer and the recently found eighth bacteriochlorophyll (BChl) molecule into account. The coupling to the non-Markovian environment is treated with a master equation derived from non-Markovian quantum state diffusion. When the excited-state dynamics is initialized at site eight, which is believed to play an important role in receiving excitation from the main light harvesting antenna, we see a slow exponential-like decay of the excitation. This is in contrast with the oscillations and a relatively fast transfer that usually occurs when initialization at sites 1 or 6 is considered. We show that different sets of electronic transition energies can lead to large differences in the transfer dynamics and may cause additional suppression or enhancement of oscillations.
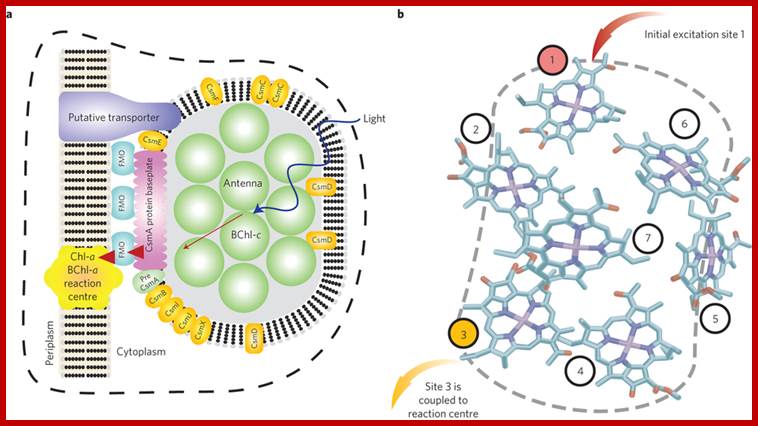
A quantum machine for efficient light-energy harvesting: The well-studied FMO complex Fenna-Matthews-Olson protein in the light-harvesting apparatus of green-sulphur bacteria exhibits some signatures of quantum coherent energy transfer. Experimental and theoretical works have scrutinized the precise mechanisms and quantumness of the energy transduction through these proteins. Research in this field might reveal new quantum mechanical principles for improving the efficiency of energy harvesting in biology. a, Diagram of the photosynthetic apparatus of green sulphur bacteria, including its antenna, energy-conducting baseplate and FMO complexes, and reaction center. The chlorosome antenna (green discs) is composed of roughly 200,000 BChl-c molecules, and is an exceptionally large structure that is designed to capture as many photons as possible in the low-light conditions the bacteria thrive in. Sunlight creates an excitation in this antenna that is transferred (red arrows) to the reaction center through one of several FMO complexes. b, The BChl-a arrangements of one of the FMO pigment-protein complexes through X-ray diffraction. The FMO complex comprises eight (although only seven are shown here) bacteriochlorophyll-a (BChl-a) molecules that are encased in a protein scaffolding (not shown). The excitation arrives from the chlorosome at one of the sites, typically thought to be the site denoted as 1. This excitation is then transported from one BChl molecule to the next. Once it arrives at site 3 it can irreversibly enter the reaction Centre and start a charge-separation process. Neill Lambert, Nature.com
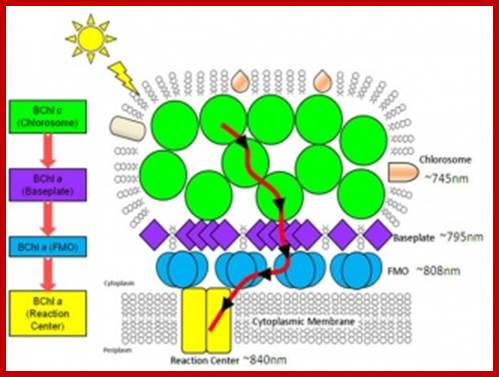
Energy transfer pathway from green bacterial chlorosomes the small structures in bacteria that absorbs light; http://www.energyfrontier.us/
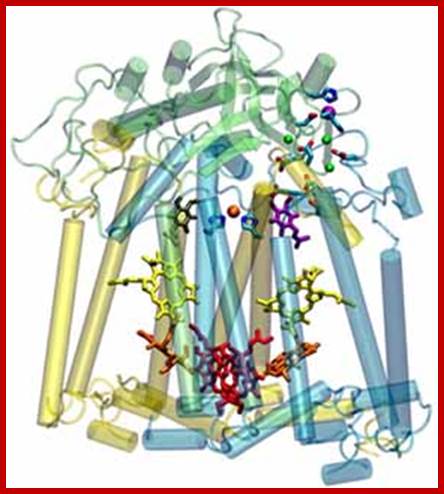
The reaction center from the photosynthetic bacterium Rhodobacter spheroids (the membrane plane is horizontal). The quasi-two-fold symmetry axis, which runs vertically, creates pairs of identical cofactors with very different properties. Examination of the protein-cofactor interactions provides insight to how Nature works at the atomic level; https://mcb.illinois.edu
Inheritance: Like Mitochondria, plastids are also inherited from one of the two parents. Mostly from mother side means egg and few from pollen, it means from male side; it is uniparental. Uniparental Inheritance is not universal.
Chloroplasts:
Chloroplasts are green colored plastids for they contain greater amounts of chlorophyll pigments. They are ubiquitously present in green plants and they are mainly responsible for providing food for themselves and to other animals in the world. These structures are mostly restricted to photosynthetic parts of the plants.
Number and Shapes:
In lower plants, such as Chlamydomonas, spirogyra, diatoms, Hdrodictyon, etc, the number of plastids present in each cell is always constant and characteristic; so also their shapes. Chlorella cells contains a single plate shaped chloroplast; where as Chlamydomonas possesses only one cup shaped chloroplast. One the other hand chloroplasts in spirogyra are ribbon shaped spirally coiled. But Zygnema cells contain two star shapes chloroplasts. In Hydrodictyon and cladophora chloroplasts are highly diffused and this is in the form of a network. In all the above said plants, plastids contain pyrenoids.
In higher land plants, the number of chloroplasts varies from cell to cell and from organ to organ, i.e. 30-200 per cell and most of them are nearly spherical or ovoidal in shape.
Distribution:
In their disposition within the cell they show some variations. In certain algae like Caulerpa and Vaucheria, chloroplasts are generally clustered in the region of the cell where it is exposed to sunlight. Even in higher plants such as tropical grasses etc, chloroplasts found in bundle sheath cells and those found in the surrounding mesophyll cells are crowded towards each other. Generally the position of chloroplasts in photosynthetic cells is not fixed, because they are constantly swept along with the protoplasmic movement.
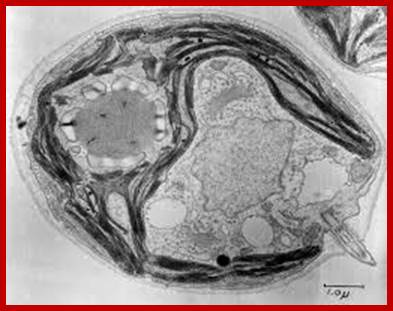
Chlamydomonas; www.cell.imagelibrary.org
Size of eukaryotic chloroplasts is 4-5 um in size but size may vary from plant to plant. Plants growing in shade contain large sized chloroplasts in their cells than that of growing in intense light.
Movement of Plastids within plant cells:
Plastid not like stationary structures, but they are dynamic structures and move in cytoplasm. Stromules are extended tubules from the surface of plastids and often interconnect plastids. Stromules are stroma- filled tubules). Actin related Myosin XI is involved in the formation and movement of stromules. There are many myosin families, but only three of them are involved in plastid movements. Chloroplasts are associated microfilaments with Myosin XI involved in vesicle and plastid movements.
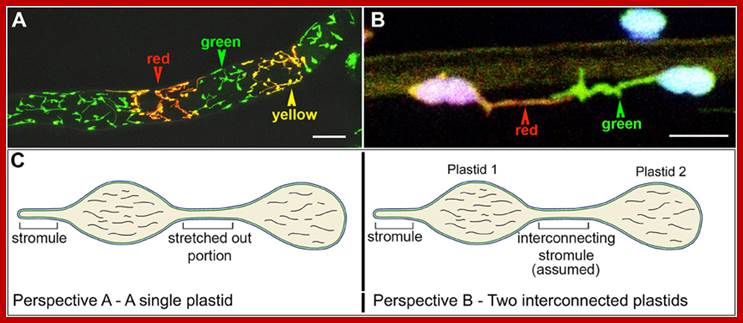
Interconnecting plastid membrane extensions; http://www.frontiersin.org/
Multiplication of plastids:
Proplastids are constantly dividing in meristematic cells and keep pace with cell division. Once proplastids develop into fully developed chloroplasts, in higher plants, they rarely divide. But in spirogyra and many algae fully developed plastids, divide at the time of cell division and they are equally distributed among the daughter cells. Stromules are found mostly in non-green plastid. In Chlamydomonas, chloroplasts divide into two to four or more at the time of reproduction. The mode of division is typical of bacterial cleavage. The time taken for such division is should take 10-18 hours. In all developing organs or plant structures plastids keep on multiplying. plastid interactions with mitochondria, the nucleus, the endoplasmic reticulum and F-actin and suggests integral roles of plastids in retrograde signaling, cell to cell communication as well as plant-pathogen interactions. Formation of stromules is active during daytime and the same is retracted during nights. And sugar induces the development of stromules. Stromules are chloroplast protrusions. Chloroplast protrusions CP appear as broad or long, grana-free extensions and occasionally form pocket-like structures with mitochondria and microbody aggregates. A loose ER cage around the plastid body, and stromules co-aligned with ER tubules were observed. The organelle interactivity suggested by these observations was attributed to the presence of membrane contact sites (MCS) between the plastid envelope and the ER.
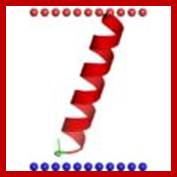
The Z-ring forms from smaller subunits of Fts-Z filaments. These filaments may pull on each other and tighten to divide the cell; http://en.wikipedia.org/
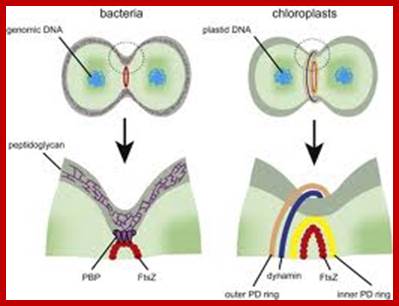
FtsZ is a protein encoded by the ftsZ gene that assembles into a ring like structure at the future site of the septum of bacterial cell division. This is a prokaryotic homologue to the eukaryotic protein tubulin. http://www.riken.jp
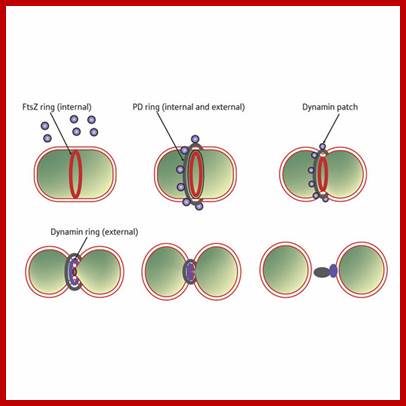
Sequence of events in chloroplast division (from top left to bottom right). An FtsZ ring forms inside the chloroplast double membrane at the division site. A plastid division (PD) ring forms outside the chloroplast double membrane at the division site. During division, dynamin patches are recruited to the PD ring. A continuous dynamin ring forms at a late stage of division. The FtsZ ring disassembles just before completion of division. When division is complete, the remnant of the PD ring and the dynamin ring disassembles outside of the chloroplast.PDV- PDVPlastid division protein http://www.riken.jp/
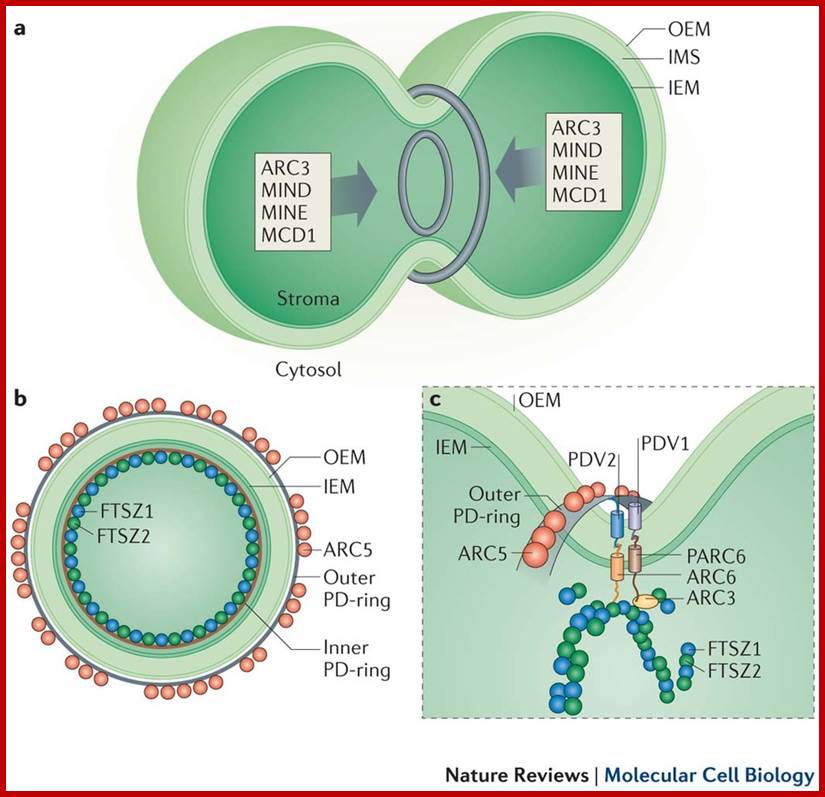
a | Plastids typically divide by binary fission. This process is mediated by concentric rings of a division machinery on both sides of the envelope (at the stromal and cytosolic surfaces of organelle). Correct positioning of the division machinery at the constriction zone is mediated by the MIN (MINICELL) system (grey arrows), which acts to control Z-ring formation. b | Multiple rings surround the organelle at the division site to enable constriction. On the stromal side, the Z-ring (comprising functionally distinct FTSZ1 (FILAMENTOUS TEMPERATURE SENSITIVE Z1) and FTSZ2 homologues) and the inner PD-ring (of uncertain composition) are present. On the cytosolic side, an outer PD-ring that is composed of polyglucan filaments and a discontinuous ring of dynamin-related ARC5 (ACCUMULATION AND REPLICATION OF CHLOROPLASTS 5) operate. c | Positional information from the stromal Z-ring is conveyed to the cytosolic components through ARC6 and PARC6 in the inner membrane and PDV1 (PLASTIC DIVISION 1) and PDV2 in the outer membrane through specific interactions in the intermembrane space (IMS). Correctly positioned PDV proteins enable the recruitment of ARC5 at the constriction zone. Z-ring formation is dynamically controlled by the action of PARC6 and ARC3. IEM, inner envelope membrane; MCD1, MULTIPLE CHLOROPLAST DIVISION SITE 1; OEM, outer envelope membrane. The images are adapted, with permission, from Ref. 107 © (2013) Elsevier and Ref. 156 © (2008) Elsevier.
![Most chloroplasts in plant cells, and all chloroplasts in algae arise from chloroplast division.[130] Picture references,[108][134]](Plant_Cellular_Structures10-Plastids_files/image016.gif)
Structure:
Al green plastids, for that matter every kind of plastids are bounded by two unit membranes; i.e. outer and inner of 7 nm thick membranes and they are separated by periplastid space of 8-10 nm thick. Unlike mitochondria, the inner membrane of fully developed plastids does not show any inward foldings; but it plays active role during the development proplastids into mature plastids. Chloroplast is filled with a liquid called stroma, in which highly organized membrane structures are found they are called grana. Besides grana, the stromatic fluid contains a host of enzymes, plastid DNA, RNAs and 70s ribosomes.
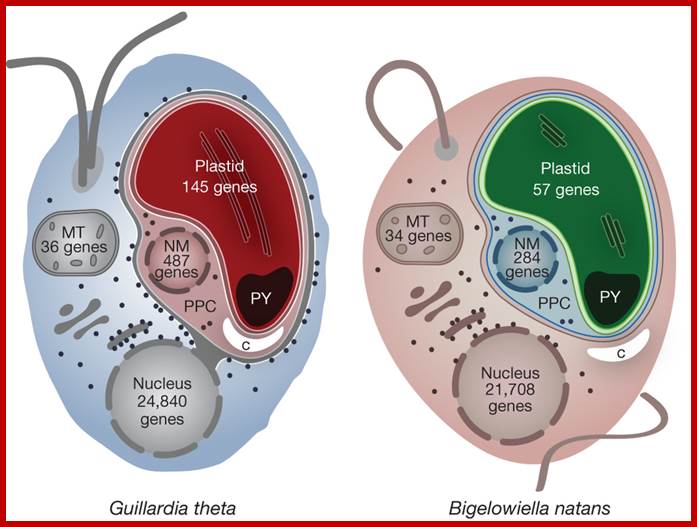
The cryptophyte alga G. theta and the chlorarachniophyte alga B.natans have plastids bound by four membranes. In cryptophytes, the outermost plastid membrane is continuous with the nuclear envelope and its surface is studded with ribosomes, which co-translationally insert nucleus-encoded, organelle-targeted proteins. Between the inner and outer membrane pairs is the periplastidial compartment (PPC), which contains the nucleomorph (NM), the relict nucleus of the eukaryotic endosymbiont. The predicted numbers of protein-coding genes in the plastid, mitochondrial (MT), nucleomorph and nuclear genomes of G. theta and B. natans are shown. Additional abbreviations: C, carbohydrate; PY, pyrenoid; http://www.nature.com/
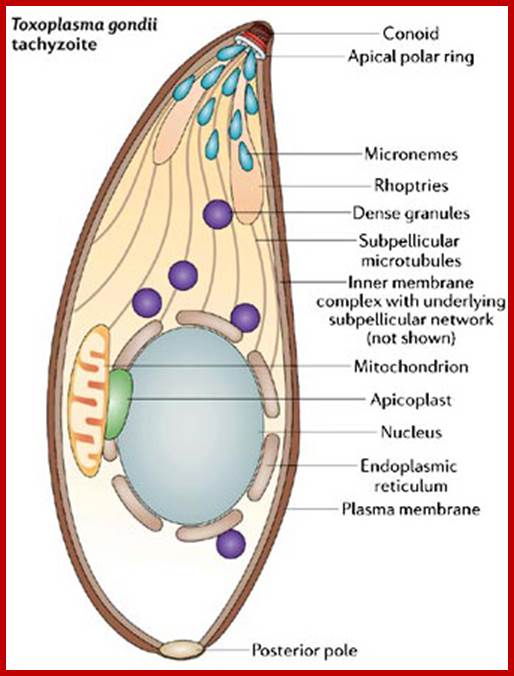
In the 1960s, microbiologists were using light and electron microscopes to study the cellular structure of apicomplexans. They discovered organelle-–like structures, different from mitochondria, first in Eimeria, and later in the Plasmodium species responsible of avian malaria (Scholtyseck & Piekarski 1965, Kilejian 1975). www.nature.com.
Regulation of apicomplexans actin based motility. Apicoplasts puzzle is why apicomplexans retained a vestigial plastid despite losing the capability for photosynthesis. A probable answer to this question is that the plastid provides a function that is important to the parasite's survival. In algae and plants, plastids are not only involved in photosynthesis, but they are also responsible for other functions. Scientists learned that apicoplasts, like some plant plastids, participate in lipid biosynthesis and iron metabolism. The analysis of both the genomic sequence and the organization of the 35-kb apicoplasts genome led to two main conclusions. The first was that the apicoplasts genome encodes enough transfer RNAs (tRNAs) and ribosomal genes for a minimal, but sufficient, translation of the protein-encoding genes present in the element. The second conclusion was that the genomic organization showed a close relationship to algae (Wilson et al. 1996). In 2008, in an exciting twist in the story, scientists studied a minute marine alga that provided evidence that apicomplexans parasites evolved from photosynthetic algal ancestors (Keeling 2008). http://www.nature.com/
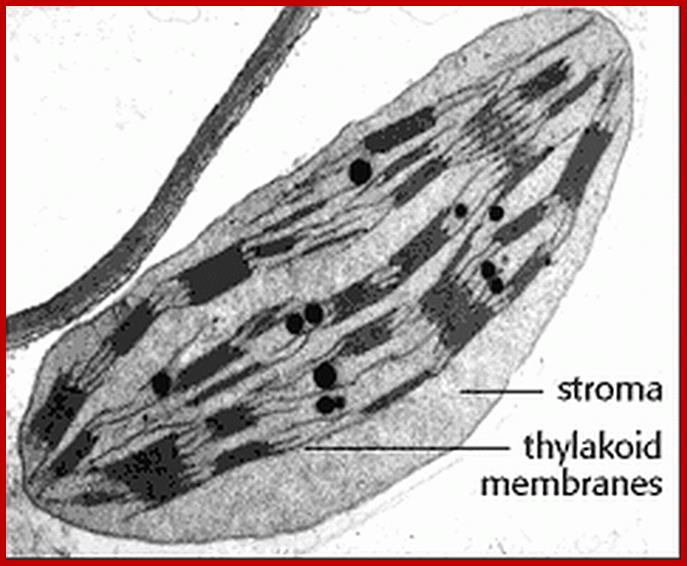
http://www.biology.arizona.edu/

Dimorphic plastid structures in sugarcane- Left bundle sheath plastids starch storing chloroplast-without granal formation, but in the right mesophyll cells one finds well developed granal differentiation; https://www.jstor.org

Grana with thylakoids
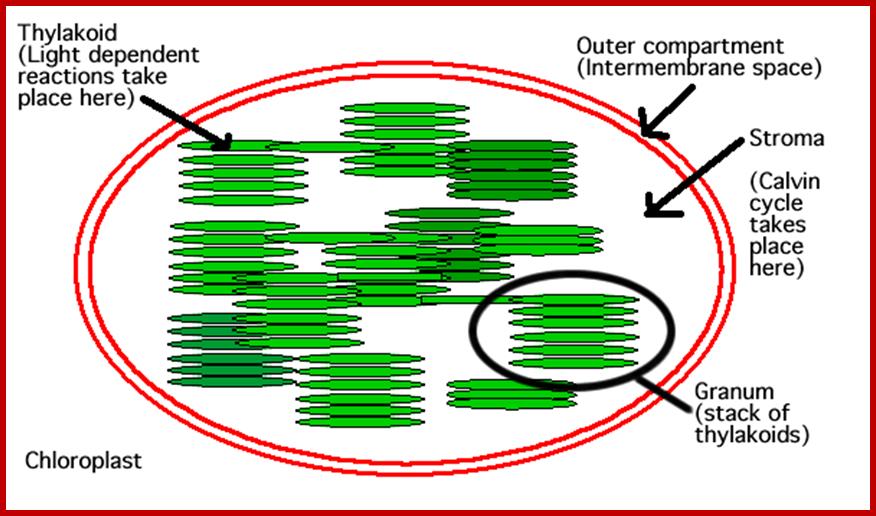
Chloroplast with grana and intergranal lamellae; www.wesharepics.info
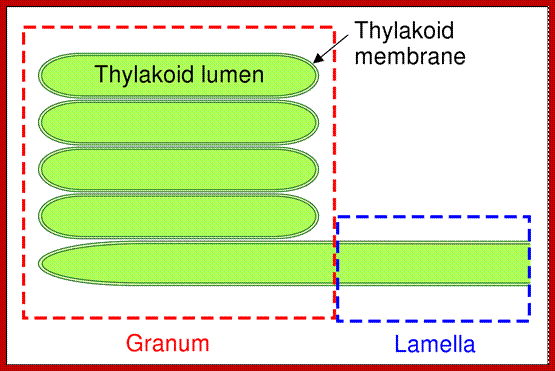
Thylakoid structure;https://en.wikipedia.org

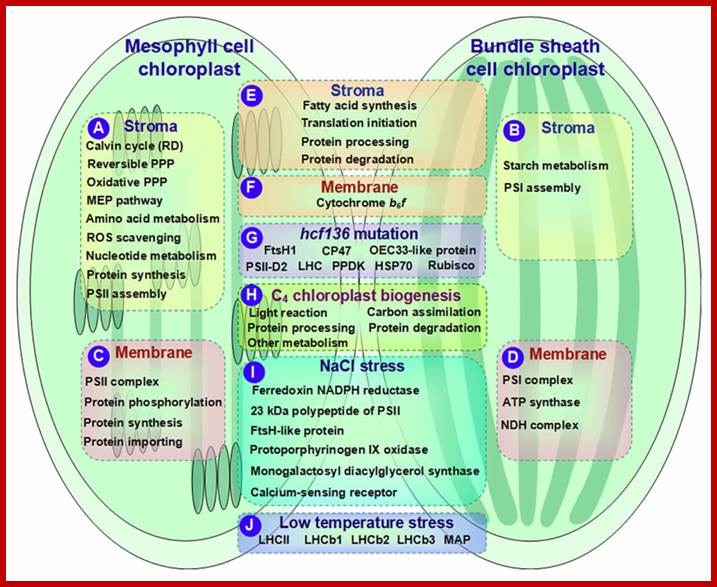
One can observe the granal organization in bundle sheath cells above and mesophyll cells below; http://biology-themiracleoflife.blogspot.in/
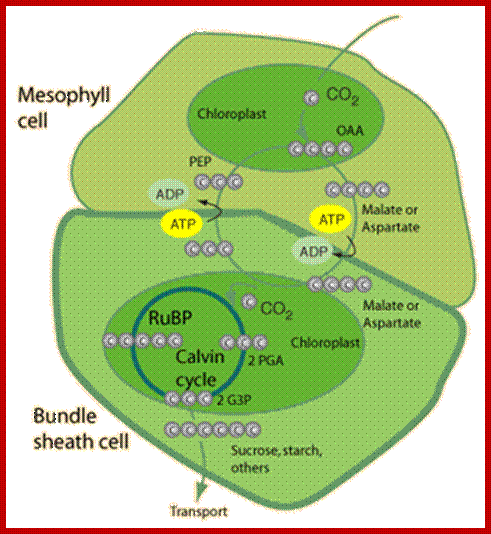
Metabolic pathways are shown to occur in Bundle sheath and Mesophyll cells; http://journal.frontiersin.org/
http://biology-themiracleoflife.blogspot.in/
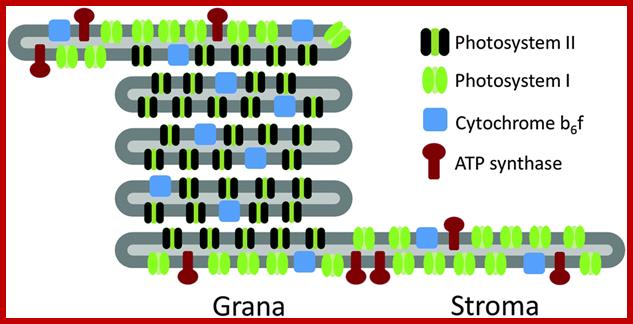
Grana:
Grana are highly specialized circular membrane sacs piled one upon another. Each chloroplast may contain 20-60 such granal clusters, a single granum can be compared to a flat circular membranous discs placed one above the other, the membrane discs are called thylakoids stacked one above the other. Such 20-60 thylakoids together constitute a granum. Moreover the grana are interconnected by another membrane structure called intergranal lamellae or stromal lamellae, which is located in between the stacks of thylakoids, but it extends from one of the thylakoid laterally, so as to form a kind of network of interconnecting membranes.
Though the above described structure holds good for most of the chloroplasts, but certain plants belonging to tropical grass members like sugarcane, zea mays, sorghum, crab grass and even some dicots like amaranthus contain chloroplasts of two types. Chloroplasts found in mesophyll cells have the same granal organization as described above, but lack in thylakoids. In its place only stromal lamellae are found. Such dimorphic chloroplasts though found in the same leaf and exhibit different functions.

Granum structure In the helical thylakoid model, grana consists of a stack of flattened circular granal thylakoids that resemble pancakes. Each granum can contain anywhere from two to a hundred thylakoids,[62] though grana with 10–20 thylakoids are most common.[76] Wrapped around the grana are helicoid stromal thylakoids, also known as frets or lamellar thylakoids. The helices ascend at an angle of 20–25°, connecting to each granal thylakoid at a bridge-like slit junction. The helicoids may extend as large sheets that link multiple grana, or narrow to tube-like bridges between grana.[77] While different parts of the thylakoid system contain different membrane proteins, the thylakoid membranes are continuous and the thylakoid space they enclose form a single continuous labyrinth. The prevailing model for granal structure is a stack of granal thylakoids linked by helical stromal thylakoids that wrap around the grana stacks and form large sheets that connect different grana; http://en.wikipedia.org/

The electron micrograph on the left (courtesy of Kenneth R. Miller and Dr.L.K.Shmway) shows the inner surface of a thylakoid membrane. Each particle may represent one photosystem II complex. In the functioning chloroplast, these particles may not be as highly ordered as seen here; Photosystems as granules located in granal membranes; www.users.rcn.com
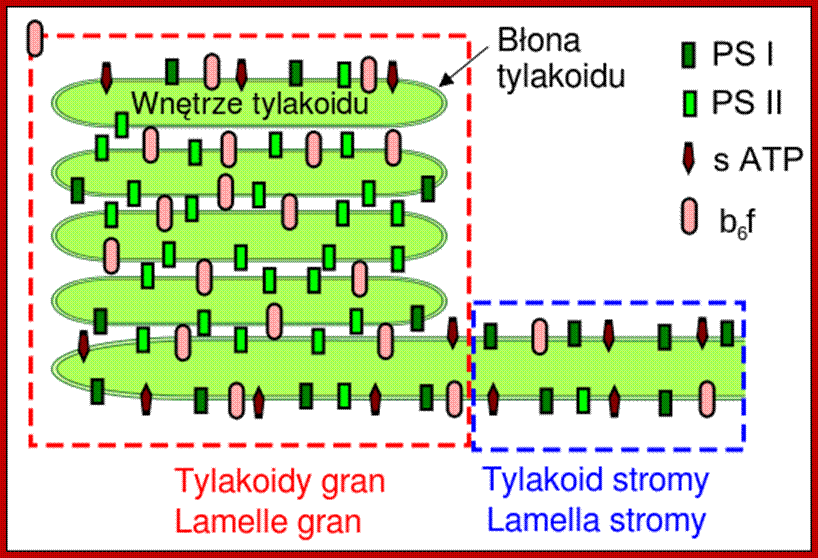
Thylakoid structural feature with Photosystems I and II and ATP synthases and in between Cyt b/f complexes.; http://www.chm.bris.ac.uk/
Thylakoid direct photobioelectro catalysis: utilizing stroma thylakoids to improve bio-solar cell performance; photo electrochemical experiments showed that stroma thylakoid electrodes generate photocurrents more than four times larger than grana thylakoids (51 ± 4 nA cm−2 compared to 11 ± 1 nA cm−2). A similar trend was seen in a bio-solar cell configuration with stroma thylakoids giving almost twice the current (19 ± 3 μA cm−2) as grana thylakoids (11 ± 2 μA cm−2) with no change in open circuit voltage; http://pubs.rsc.org/
Chemical Composition:
Chloroplast limiting membranes contain normal lipid and protein components. But the granal membranes including both thylakoid and intergranal membranes, posses 20-30% of the lipids and the rest of it is all proteins. The most common lipids found in these membranes are ethanolamine, sulfolipids, phytosterols and glycolipids and pigments. The stromal fluid contains enzymes and their required components for Dark reactions and even lipids particularly fatty acids are produced
Pigments:
Plant pigments are basically simple lipids containing many isoprenoid units. Association with other compounds brings about variations in them. Among the plant pigments present in plastids, chlorophylls and carotinoids are found in 3:1 proportions. The composition of pigments varies significantly among different groups of plant kingdom. Chl a, Chl b, β-carotene and xanthophylls are found in most of the green plant chloroplasts. But lower plants contain diverse pigments composition. Nonetheless, Chl- a is found in all photosynthetic plants. In fact, it acts as a primary pigment and all others including phycoerythrin and phycocyanin are considered as accessory pigments. Different kinds of pigments can be identified by their absorption spectrum.
Chlorophylls:
Chlorophyll pigments are unique molecules of the biological world. They are made up of hydrophilic head and a hydrophobic tail. The head consists of four pyrole rings joined to each other by a single master ring of CH bridges. The inorganic Mg2+ ion is found in the centre of the ring. If Mg2+ is replaced with Fe2+, this ring becomes a heme group found in animal hemoglobin. However, chlorophyll also contains an additional long chain of saturated hydrocarbons called phytol chain. But Chl a is distinguished from Chl b in having CH3 group in II ring, while Chl b possesses CHO. Pheophytin is similar to chlorophyll, but lacks Mg2+.
Carotinoids:
Carotinoids are simple lipids containing long chain of isoprenoid hydrocarbons. They are soluble in lipids. There are of two types of carotinoids namely carotenes and xanthophylls. Carotene pigments are made up of short chains of unsaturated hydrocarbons with hexane rings at each end. Xanthophylls also have similar hydrocarbon chains with hexane rings, but contain quite a number of hydroxyl groups.
Proteins:
Analysis of granal membrane proteins on SDS slab gel-electrophoresis reveals the presence of more than 200 polypeptides. A most of them are structural proteins, light harvesting antenna proteins, electron transporting proteins. Oxygen releasing Z-proteins, along with them ATP synthase and RUBP carboxylase are also found associated with granal membranes. Majority of them are imported from cytosol
Light harvesting proteins are a group of proteins that are associated with various pigments and they are responsible for harvesting and transferring solar energy. The mol wt of these proteins ranges from 11 KD to 46 KD. The core LHP proteins however has mol wt. of 11-16 KD and the peripheral LHP proteins have a mol. Wt. of 24-46 KD. Most of the proteins exhibit hydrophilic as well as intrinsic hydrophobic properties.
The oxygen releasing Z-protein is found associated with photosystem II and it has a mol wt. of 64 kd. It is made up of two subunits of 32 KD each. The Z-protein is associated with 4 Mn2+ ions, which are tightly bound to the protein moiety. The Z-mn2+ protein complex is a hydrophobic protein and it is mainly responsible for oxygen liberation. Similar to the above mentioned protein complexes, electron transporting proteins, ferrodoxin reducing proteins, NADP reductase proteins and others are grouped into complexes. The cyt.b6-cyt.f protein complex has non heme iron and also possesses phospholipids and carotinoids but no chlorophylls. The mol. Wt. of this protein is 103 KD.
The enzymatic complex required for ATP synthesis and its associated coupling factor proteins are situated at the outer surfaces of thylakoids. They are half buried in the membranes in such a way that the head is exposed to stromatic fluid. This ATP synthetase complex called CF 1 has 5 subunits of mol. wt 350KD. But the protons secreting hydrophobic protein called HF (mol. Wt 32KD), has two subunits and it is buried in the core of the thylakoid membrane, at the same time it is also in contact with CF1 structure.
Another enzyme complex which is mainly responsible for the fixation of CO2 is RUBP carboxylase. Its mol. Wt is 55.8KD. It consists of 8 large subunits and 8 small subunits. This enzyme is located at the outer surface of thylakoid membranes. Interestingly RUBP carboxylase is one of the most abundant proteins found in the biological world. More than 50% per cent of the total proteins found in leaves is RUBP carboxylase.
Structural Organization of Thylakoid membranes:
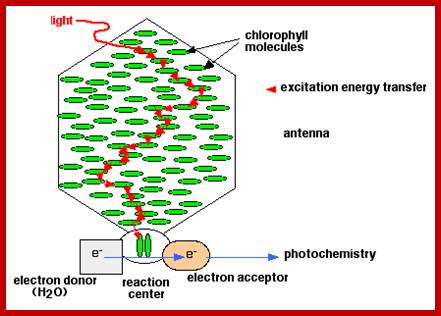
Freeze fracture – electron microscopic studies, combined with biochemical analysis of various components of granal membranes reveal, that thylakoid membrananes contain a large number of granular structures of various dimensions. Most of them are embedded in the core of membranes. The components are not fixed in a position, but exhibit lateral mobility true to the nature of dynamic fluid nature of membranes. The granular particulates were observed by Briggs and Park. Later they were isolated and purified into different fractions and chemical analysis was made. Such granular structures are called Quantosomes, because they are mainly responsible for capturing quantum of photons of solar electromagnetic radiation.
The quantosomes found in thylakoid membranes are of different sizes and dimensions but they show vectorial disposition within the membrane. The bigger quantosomes are located in the core of thylakoid lipid layers spanning the entire cross section projecting towards both stromal side and lumen side. On the other hand, smaller quantosomes are placed in the core of membrane but more towards stromatic surface in such a way ¾ of the structure is embedded in the lipid core. The granular particles of both sizes are organized in such a way each of them fit well into the spaces found in between them.
The larger quantosomes are called photosynthetic units or photosystem-II and the smaller as PS I. Nonetheless, there are other particulate which are of the same size as PS I, but have different composition and functions. However intergranal lamellar organization is quite different from that of thylakoid membranes. The intergranal lamella is almost lacking in PS II quantosomes, but contain mostly PS I systems.
The organization of quantosomes found in chloroplasts of bundle sheaths of C4 is quite different from that of other normal chloroplasts. These chloroplasts contain only stromal lamellae. Even though PS II is more or less absent from intergranal membranes, one may find few of them, but contain PSI systems.
Photosynthetic units like PS I and PS II have different chemical composition. Still each of these units are made up of 250-300 Chl molecules complexes with LHP antenna proteins and other cofactors required for specific photochemical reactions.
More details about their composition and structure have been described in the chapter photosynthesis. The role of PS I ad PS II present in thylakoid membranes is to perform NADP reduction, noncyclic photophosphorylation and oxygen liberation. On the contrary, the intergranal lamellae containing just PS I systems perform just cyclic photophosphorylation.
The structural and functional specificity applies to C4 chloroplasts also, where chloroplasts of mesophyll cells perform noncyclic photophosphorylation, NADP reduction and liberate oxygen. But the stromal lamellae found in chloroplasts of bundle sheath cells perform cyclic photophosphorylation only for they contain only PS I. system.
Stroma:
Amorphous, often semi viscous fluid present within the chloroplast membrane is called Stroma. A large number of enzymes responsible for carbon fixation, amino acid synthesis, protein synthesis, nucleic acid metabolism, pigment synthesis, N2 metabolism and fatty acid synthesis are present in stromatic fluid. It has also enzymes for the synthesis of Gibberellic acid and Abscisic acid. Some of the biosynthetic pathways in the stroma are under the control of various factors like light, phytochromes, temperature and photoperiods. Though chloroplasts have their own genetic material, the synthesis of various components required for chloroplast is under the dual control of nuclear genome and plastogenome.
In the case of C4 plants, the stromatic fluid of chloroplasts found in mesophyll cells contain enzymes for Hatch and Slack pathway. Such enzymes are totally absent in the stroma of C3 chloroplasts. On the other hand, C4 chloroplasts found in bundle sheath cells possess enzymes for C3 pathway and also contain malate dehydrogenase. Thus C4 and C3 chloroplasts show dimorphism both in their structure and in function.
Another important feature of chloroplasts is the presence of circular DNA, various species of tRNAs and 70s ribosomes. Having its own genetic material and translation machinery chloroplasts enjoy semiautonomous state in the cell, a feature similar to that of mitochondria.
Chloroplast DNA:
Plastid DNAs are circular duplex molecules with a total length of 45 mm. But in some cases DNA of 15 mm have been isolated. However each plastid consists of 6-30 copies of circular DNAs and most of them are in super coiled state. Based on its genomic size, it has been calculated that each cp DNA molecule can code for about 110-120 proteins. Chloroplast DNA when subjected to ultra centrifugation, settles as satellite DNA, because of high GC content.
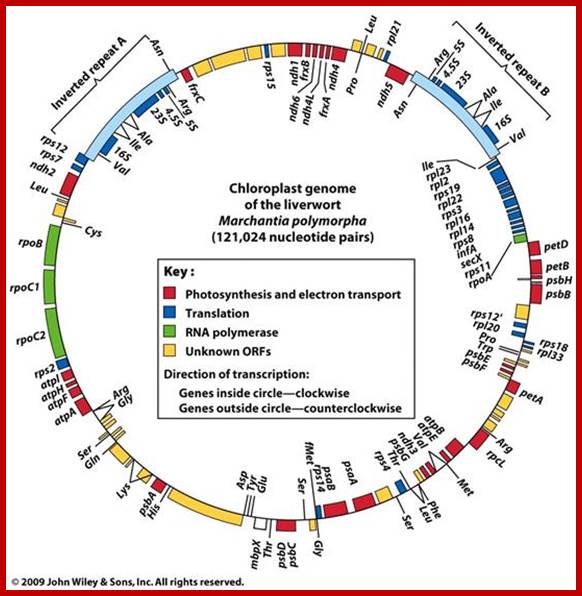
Chloroplast gene organization- Marchantia polymorpha; The DNA is circular, 121,024 base pairs (bp) long; contain 128 gene; Chloroplasts contain their own autonomously replicating DNA genome. The majority of proteins present in the chloroplasts are encoded by nuclear DNA, but the rest are encoded by chloroplast DNA and synthesized by the chloroplast transcription–translation machinery1–4. Although the nucleotide sequences of many chloroplast genes from various plant species have been determined, the entire gene organization of the chloroplast genome has not yet been elucidated for any species of plants. To improve our understanding of the chloroplast gene system, we have determined the complete sequence of the chloroplast DNA from a liverwort, Marchantia polymorpha, and deduced the gene organization. As reported here the liverwort chloroplast DNA contains 121,024 base pairs (bp), consisting of a set of large inverted repeats (IRA and IRB, each of 10,058 bp) separated by a small single-copy region (SSC, 19,813 bp) and a large single-copy region (LSC, 81,095 bp). We detected 128 possible genes throughout the liverwort chloroplast genome, including coding sequences for four kinds of ribosomal RNAs, 32 species of transfer RNAs and 55 identified open reading frames (ORFs) for proteins, which are separated by short A+T-rich spacers (Fig. 1). Twenty genes (8 encoding tRNAs, 12 encoding proteins) contain introns in their coding sequences. These introns can be classified as belonging to either group I or group II, as described for mitochondria5. Interestingly, seven of the identified ORFs show high homology to unidentified reading frames (URFs) found in human mitochondria; KANJI OHYAMA et al; http://www.nature.com/
Gene mapping of chloroplast DNA has shown that it has two sets of genes for ribosomal RNAs. It also has genes for the large subunit of RUBP carboxylase, hydrogen secreting proteins, all tRNAs, all aminoacyl synthetases, RNA polymerases, DNA polymerase, some LHPs, ALA synthetase etc.
Chloroplast DNAs replicate by D-loop mechanism by a DNA polymerase coded for by its own genome. Transcription is performed by its own RNA polymerases which is sensitive to rifampicin. But plastogenome expression is controlled by light, nuclear factors and other environmental factors. Phytochrome which is also present in plastids control or assist gene expression during greening and development of plastids. Added to this, plastids are also involved in the expression certain gene products which are responsible for the synthesis of GA ad ABA under certain environmental conditions. The plastogenome also contains genes for male sterility in specific plants. Recent studies indicate plastogenome expression has a direct bearing on floral induction.
Chloroplast ribosomes are more or less similar to prokaryotes in terms of size is 70s and rRNA content. Their functional activity is also inhibited by chloramphenicol similar to that of bacteria and mitochondria.
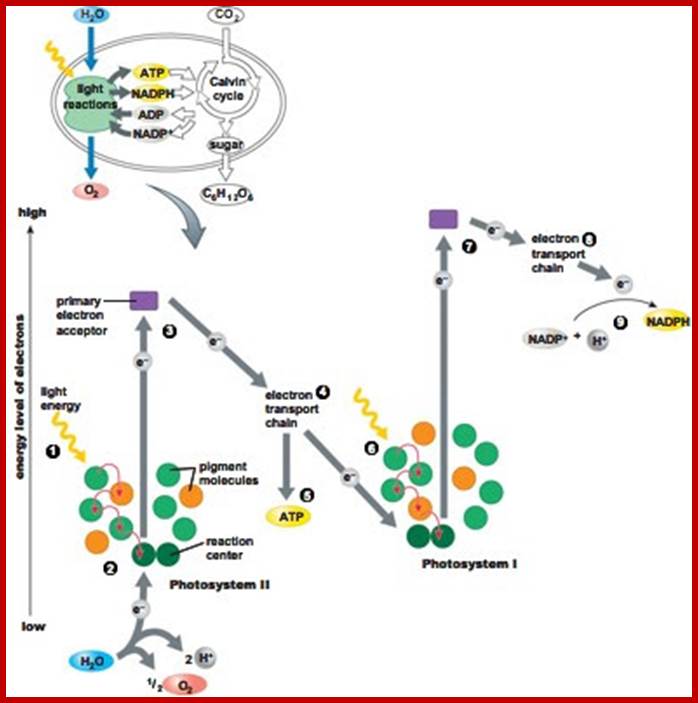
Chloroplast photosystems involved in primary light reactions:

Photosynthesis; www.uic.edu
Photosynthetic unit; www.sites.bio.indiana.edu

Cyclic Photophosphorylation- produce ATP only
http://www.uic.edu/
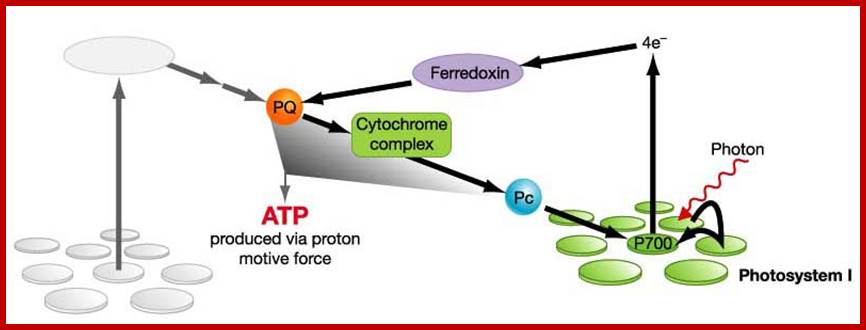
- Reaction centre for photosystem I is P700.
- 2 electrons excited by light energy, achieve high energy level, accepted by FeS (primary acceptor).
- The 2 electrons flow to/received by Ferredoxin, received by cytochrome complex, then received by plastocyanin, then received back by photosystem I.(ADP is used & ATP is produced).
- The flows of electrons provide energy to pump protons across thylakoid membrane. This activated ATP synthase to produce ATP.
Noncyclic Photophosphosphorylation:
Noncyclic Photophosphosphorylation produces NADPH and ATP; http://cascadingbooks.blogspot.in/
Reaction centre for photosystem II is P680.
- H2O is broken down to provide 2 electrons and O2.
2. 2 electrons excited by light energy and achieve high energy potential/energy level, accepted by Pheaophytin (the primary acceptor).
3. 2 electrons flow to/received by plastoquinones, received by cytochrome complex, received by plastocyanin, and received by photosystem I(ADP is used & ATP is produced).
The flow of electrons, provide the energy to pump protons across the thylakoid membrane. This activates ATP synthase to produce ATP.
From photosystem I, the reaction centre is P700.
The 2 electron excited by light energy and achieve high energy level, and accepted by FeS.
2 electron flow to/received by Ferredoxin and received by NADP reductase.
Enzyme NADP reductase converts NADP+ to NADPH and H+ ion.
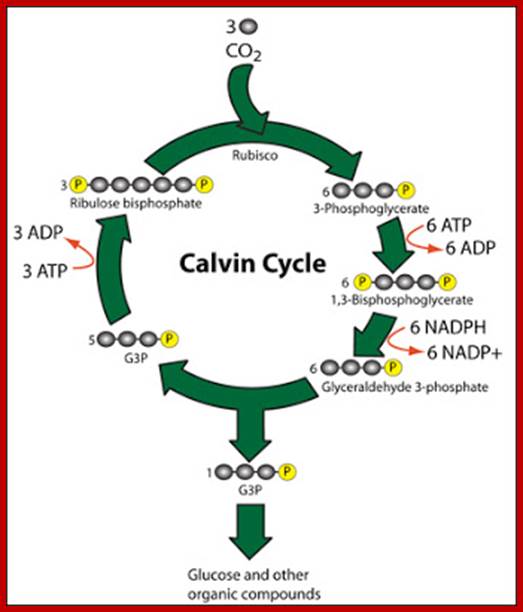
Dark Reaction:
Calvin Cycle
http://cascadingbooks.blogspot.in/
· CO2 combines with 5 carbon compound, RuBP with the help of enzyme rubisco to form unstable 6 carbon compound.------>(CARBON DIOXIDE FIXATION phase)
· 6 carbon compound split into 2 molecules of 3-phosphoglycerate/phospho-3-glycerate.
· 3-phosphoglycerate undergoes a series of reduction processes to form glycealdehyde-3-phosphate/triose phosphate. ATP & NADPH is used.------>(REDUCTION phase)
· Glyceraldehyde-3-phosphate undergoes regeneration to regenerate RuBP.------>(REGENERATION OF CO2 ACCEPTOR phase)
· Some glyceraldehyde-3-phosphate assimilate to form carbohydrates, lipids, & proteins.------>(PRODUCT SYNTHESIS phase)
·
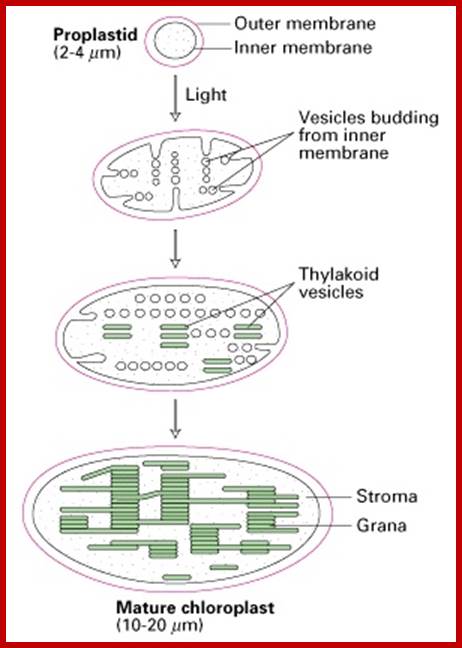
· The utility of understanding membrane biogenesis. Formation of chloroplasts from proplastids begins by the light-induced budding of the inner membrane. It is very useful to understand the morphogenesis of membrane systems and organelles, since this information can guide the researcher to evolutionarily related membrane systems in other organisms. Here we see that the thylakoids are formed from vesicles that pinch off from the inner chloroplast membrane and then fuse to form thylakoid vesicles. Consistent with the idea that chloroplasts evolved from ancestral photosynthetic bacteria, the inner membrane of the chloroplast has similarity to the bacterial plasma membrane, and the process of translocation across thylakoid membranes has similarities with the secretion of bacterial proteins across the bacterial plasma membrane, a process that has been intensively studied. Here is a review question for you: What are the characteristics of protein-secreting pathways of the bacterial plasma membrane that are similar to the translocation pathways across the thylakoid membranes? What bacterial compartment does the lumen of the thylakoid most resemble?; http://web.uconn.edu/
· 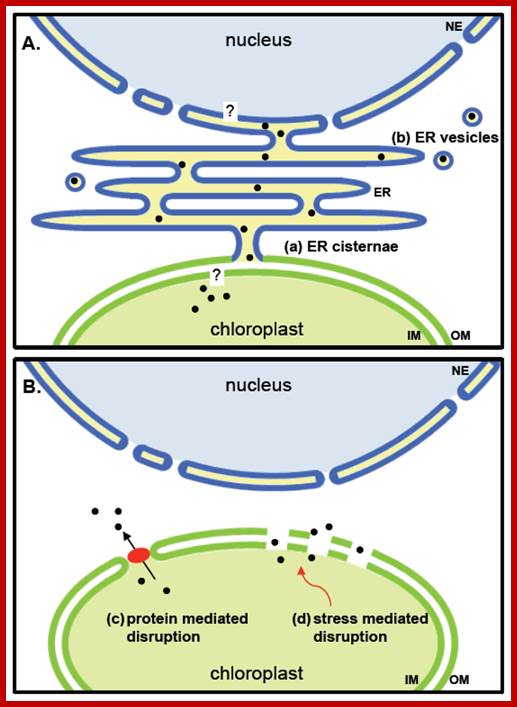
· Selected mechanisms of protein translocation from chloroplasts to the nucleus. (A) Putative ER mediated transfer of plastid proteins to the nucleus. A periplasmic space formed by ER cisternae and intermembrane space of plastids has been observed under certain conditions (see text). Proteins from the stroma of plastids would need to transverse a single membrane to become included in this space, which is continuous with the envelope of nuclei. To enter the nucleus, proteins would need to cross the inner membrane of the nuclear envelope; (B) Hypothetical release of proteins directly into the cytoplasm. Transient pores might be formed by activity of proteins such as TGD2 being involved in lipid exchange between ER and plastids [90]. Small disruptions in the membrane leading to a leakiness of chloroplasts might occur upon stress. Black dots, released plastidic proteins; red ellipse, protein complex which mediates membrane permeability; NE, nuclear envelope; ER, endoplasmic reticulum; IM, inner membrane; OM, outer membrane. http://www.mdpi.com/
·
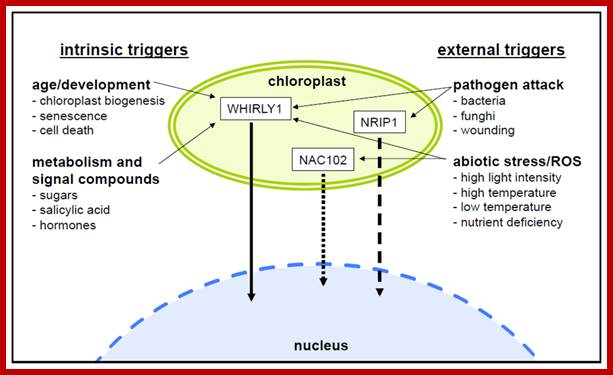
· Changes in the developmental or metabolic state of plastids can trigger profound changes in the transcript profiles of nuclear genes. Many nuclear transcription factors were shown to be controlled by signals generated in the organelles. In addition to the many different compounds for which an involvement in retrograde signaling is discussed, accumulating evidence suggests a role for proteins in plastid-to-nucleus communication. These proteins might be sequestered in the plastids before they act as transcriptional regulators in the nucleus. Indeed, several proteins exhibiting a dual localization in the plastids and the nucleus are promising candidates for such a direct signal transduction involving regulatory protein storage in the plastids. Among such proteins, the nuclear transcription factor WHIRLY1 stands out as being the only protein for which an export from plastids and translocation to the nucleus has been experimentally demonstrated. Other proteins, however, strongly support the notion that this pathway might be more common than currently believed. http://www.mdpi.com/
· Fate of chloroplast products
·
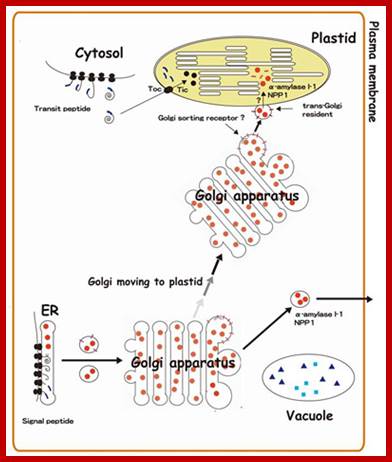
· Golgi-to-Plastid Communication Operates in Plastid Targeting of AmyI-1 and NPP1;
· Protein targeting into plastids is an essential cellular event for expressing plant function and maintaining plant life. Most of chloroplast proteins are known to be synthesized in the cytosol as precursors with an N-terminal transit peptide and imported post-translationally through the Toc-Tic complex into the organelle. Recently, we found that glycoproteins, AmyI-1 and NPP1 are occurred in plastids of rice living cells and play significant roles in the starch metabolism in plastids. We propose a novel pathway for targeting glycoproteins from the endomembrane system into the plastids (Fig. 3), which involves direct communication between the Golgi apparatus and the plastids. http://www.agr.niigata-u.ac.jp/
·
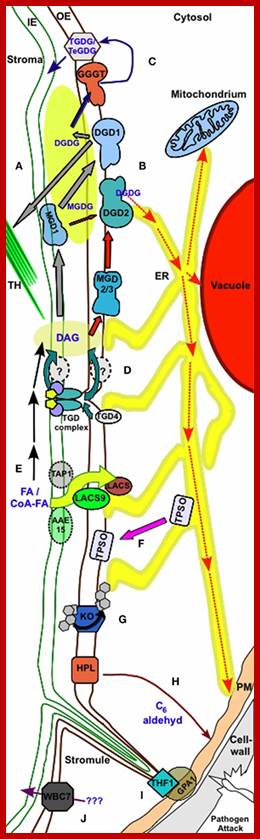
· The G family ABC transporterWBC7 mediate transport of unknown compounds through the OE. Process in the outer Plastid envelop; http://journal.frontiersin.org/
Stromules:
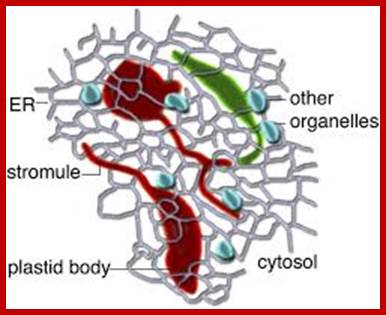
A Diagrammatic Representation of the Possible Role of Plastid Stromules. As extensions from independent plastids (red or green), stromules likely allow more effective communication and interaction with other cellular components, such as the endoplasmic reticulum (ER); small organelles, such as mitochondria and peroxisomes (depicted in blue); and the cytosol in general. Martin H. Schattat Martin-Luther-U; http://www.plantcell.org/
Chloroplast Signaling:
Chloroplast regulate intercellular trafficking through plasmodesmata; it means they regulate al aspects of plant growth and development by transporting metabolites, hormones, transcription factors and small RNA molecules. Chloroplast signaling;. in fact plastid derived signals regulated nuclear gene expression. Products of proteins, lipids and RNA oxidation products act as signals and regulators of gene expression in plant systems.
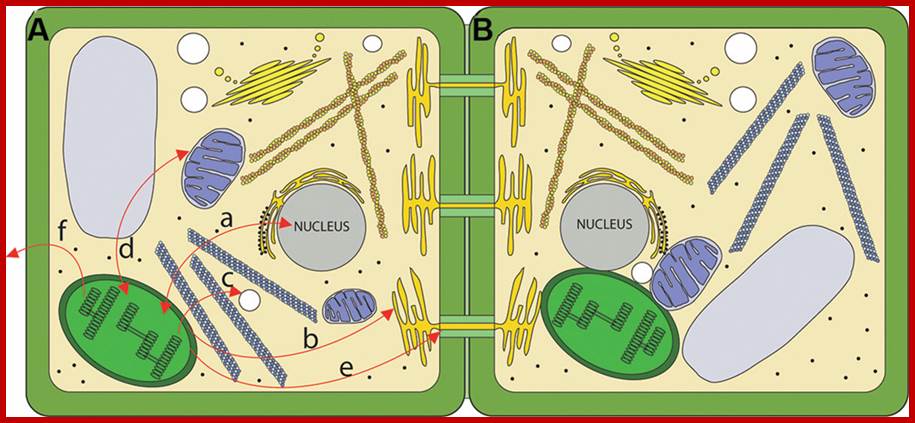
Routes for chloroplast signaling. (A) Chloroplasts generate signals that target multiple intercellular targets. (a) The majority of chloroplast proteins are encoded by the nucleus, and the import of those proteins into the chloroplast is anterograde signaling. In turn, several chloroplast products act as retrograde signals to regulate expression of nucleus-encoded genes. (b) Chloroplasts are metabolically coupled to the ER and it is likely that signals may move from the chloroplast to the ER. (c) Chloroplasts and peroxisomes are also closely associated, and numerous chloroplast products are substrates for peroxisomal pathways. (d) Mitochondria and chloroplasts are known to signal to each other. (e) Chloroplast signals regulate intercellular trafficking via plasmodesmata. It is not clear if this signaling is direct or involves retrograde signaling to the nucleus. (f) Chloroplasts produce volatile compounds that can signal to neighboring plants during pathogen attack. (B) The physical interaction between chloroplasts and various organelles may serve as a direct route for signaling. http://journal.frontiersin.org/
Biogenesis:
Plastids are generally inherited through maternal side as in the case of mitochondria. During the development of plant structures, proplastids multiply and they are evenly or unevenly distributed among daughter cells. Every cell in the plant body possesses plastids. The pattern of inheritance itself indicates that plastids are derived from pre existing plastids.
To begin with, proplastids contain a paracrystalline lattice in stroma. With the onset of light as the stimulus (red light is enough), the gene for aminoleuvulinic acid synthetase is derepressed, in the sense it is activated. The product of gene expression is ALA synthetase. The production of this enzyme can be inhibited by CAP but not with CHI, which suggests that they are light sensitive late gene products, which in turn activate light insensitive constitutive genes. Though the development of chloroplasts is interdependent of nuclear genes and plastogenome expression at certain stages of development, chloroplasts show autonomy. For example, the division of proplastids in Euglena is independent of cell division, but further development requires the products of both nuclear genes and plastogenes.
When proplastids are exposed to light they gradually turn green and enlarge in size. This is accompanied with the development of granal structures. During these stages, the inner chloroplast membrane produces a number of finger shaped invaginations. They in turn pinch off a number of membranous vesicles, which accumulate in the centre. The vesicles start fusing with one another and finally organize into clusters of thylakoid membranes called Grana. Once the development of chloroplast is completed the invaginations of inner membranes disappear.
Plastids by virtue of having its own genetic material and ribosomal translating machinery exhibits semi autonomous state within the cells. The inheritances pattern also demonstrates the same. Inheritance of chloroplasts is maternal and non Mendelian cytoplasmic type. The Mendelian pattern is controlled by nuclear genome, but the cytoplasmic inheritance is controlled by plastogenome. Though plastids have their own genome, they need the co-ordination of nuclear genes and its products for the completion of the development of chloroplasts.
Such interactions between plastogenome and nuclear genome can be observed during the development of proplastids into green chloroplasts. It is very well known that chloramphenicol, an antibiotic, inhibits the translation of 70s ribosome mediated protein synthesis. On the other hand cycloheximide inhibits cytosolic 80s ribosome mediated protein synthesis. If CAP is added during the greening of proplastids, pigments continue to accumulate in the thylakoid membranes, but electron transport activity is inhibited. In addition to this the membranes vesicles generated by inner plastid membrane do into fuse with one another to form thylakoid membranes. On the contrary, if CHI is added during the development of proplastids, greening is inhibited but the thylakoid membrane formation takes place partially (50%). The above results suggest but there is an interaction between the nuclear genome and plastogenome products in the biogenesis of thylakoids and its components.
Studies in this regard show that a large number of protein complexes found in chloroplasts are found to be nuclear gene products and 120 or so proteins are coded for by the plastogenome. For example, ferrodoxin and plastoquinones associated proteins, 32KD protein of photosystem II, some of the LHP proteins for Chl. A/b of PS II small subunit protein part of RUB carboxylase is coded for by the nuclear genome. On the other hand the production of small subunits of RUBP carboxylase which is essential for the expression of large RUBP subunit is the product of nuclear gene. Another interesting observation is that one of nuclear gene product affects the binding of RNA polymerase to plastid DNA. The RNA polymerase itself is a product of plastogenome. However there are no clear cut reports to show that plastogenome products control the nuclear gene expression required for plastid development.
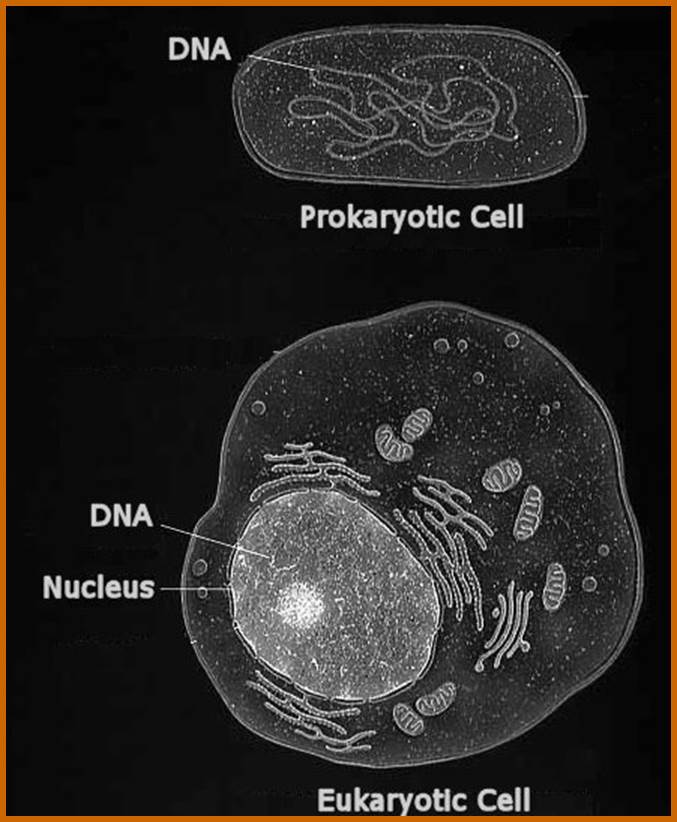
The first eukaryotic cell evolved more than a billion years ago. Since then, these organelles have become completely dependent on their host cells. For example, many of the key proteins needed by the mitochondrion are imported from the rest of the cell. Sometime during their long-standing relationship, the genes that code for these proteins were transferred from the mitochondrion to its host's genome. Scientists consider this mixing of genomes to be the irreversible step at which the two independent organisms become a single individual. http://www.fossilmuseum.net/
Origin Mitochondria and Plastids in Eukaryotic Plants:
Eukaryotes are those which contained Nucleus without mitochondria One such organism or like, is Monocercomonoides found in the guts of Chinchella. Micromonoides did not show any trace any mitochondrial required components. Their genome sequence revealed no signs of mitochondrial genes in the microbe. Further examination also showed that Monocercomonoides are lacking all the key proteins that enable mitochondria to function. Such eukaryotes engulfed a bacterium which can utilize oxygen and generate energy. This is called primary endosymbiosis. Later the same organisms have undergone secondary endosymbiosis. Such event might have happened with Achaeans about 2500 million years ago. The first got endosymbioisized was mitochondria. It is to be remembered acquisition of a mitochondrion was an important and perhaps defining event in the evolution of eukaryotic cells,
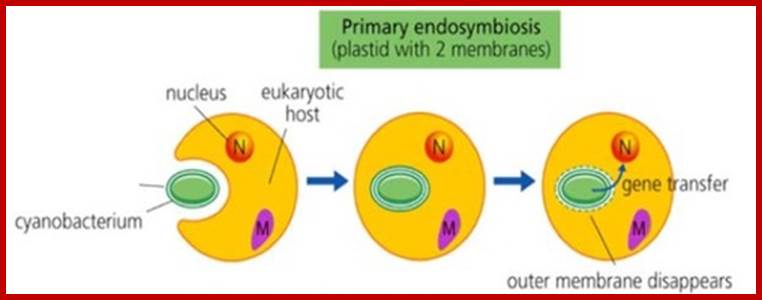
Endosymbiogenesis of mitochondria and plastids; www.lanelaburi.wordpress.com
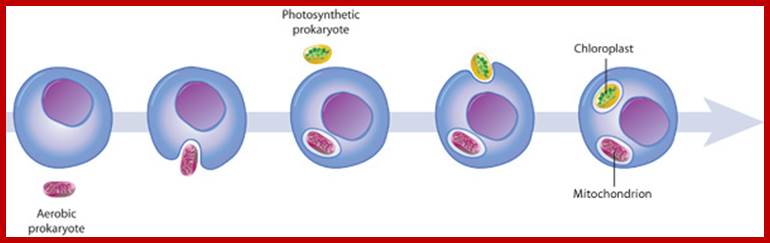
The origin of mitochondria and chloroplasts; Mitochondria and chloroplasts likely evolved from engulfed prokaryotes that once lived as independent organisms. At some point, a eukaryotic cell engulfed an aerobic prokaryote, which then formed an endosymbiotic relationship with the host eukaryote, gradually developing into a mitochondrion. Eukaryotic cells containing mitochondria then engulfed photosynthetic prokaryotes, which evolved to become specialized chloroplast organelles.; http://www.nature.com/

Secondary symbiosis led to the development of Chloroplasts after Mitochondria.
Like animal mitochondria, which are transmitted to their offsprings only through maternal parents; plastids (chloroplasts cp) like mitochondria are also inherited through maternal inheritance; ex. Mirabilis jalapa, where pollen have no plastids.
Although this maternal inheritance is often considered a general phenomenon of organelle gene inheritance, it is not. In Pelargonium zonale, Baur found that many progeny received chloroplasts from the female parent only, but a number received plastids from both parents, producing variegated progeny in which the maternal and paternal chloroplasts segregated into different cells. A few progeny showed chloroplasts of only the paternal type. Baur correctly guessed that this is a consequence of the random segregation of plastids between embryonic and extra-embryonic cells, so that embryos and adult plants sometimes received only green or only white plastids. From extensive studies it is believed that more than 80% of the Angiosperms strictly chloroplast and mitochondria are maternal. http://www.sciencedirect.com/
In Caenorhabditis elegans-Fertilization-triggered autophagy eliminates paternal mitochondria.
But In Sequoia sempervirens, in which mtDNA and cpDNA are paternally inherited. But in mirabilis jalapa cp DNA is inherited vis maternal process. In the Geranium plant (Pelargonium zonale), cpDNA is inherited maternally, paternally or biparentally . When zygotes receive cpDNA from both parents, cpDNAs from each parent are rapidly segregated following cell division, resulting in the mature plant inheriting clonal sectors of cells that are homoplastic for only 1 type of cpDNA.
In angiosperm species, such as Arabidopsis thaliana, both mtDNA and ptDNA are maternally inherited. In A. thaliana, the active degradation of paternal mtDNA and ptDNA takes place during pollen development .
The content of mtDNA and cpDNA gradually decreases during meiosis, and these DNAs are no longer detectable in mature pollen. The wild-type DPD1 encodes a pollen-specific Mg2 +-dependent exonuclease, which is transported into both chloroplasts and mitochondria, suggesting a direct role for this exonuclease in the degradation of mtDNA and ptDNA. http://www.sciencedirect.com/.
Uniparental inheritance of mitochondrial DNA in many sexually reproducing species: This is accomplished by various strategies such as 1)decrease in the content of mtDNA during spermatogenesis, 2) elimination of mtDNA from mature spermatozoa, 3) prevention of sperm mitochondria from entering the oocyte, 4) active degradation of the paternal mtDNA in the zygote, 5) selective degradation of the whole paternal mitochondria in the zygote.
The similarities between the genomes of chloroplasts and bacteria are striking. The basic regulatory sequences, such as transcription promoters and terminators, are virtually identical in the two cases. The amino acid sequences of the proteins encoded in chloroplasts are clearly recognizable as bacterial, and several clusters of genes with related functions (such as those encoding ribosomal proteins) are organized in the same way in the genomes of chloroplasts, E. coli, and cyanobacteria.
In about two-thirds of higher plants, the chloroplasts from the male parent (contained in pollen grains) do not enter the zygote, so that chloroplast as well as mitochondrial DNA is maternally inherited. In other plants, the pollen chloroplasts enter the zygote, making chloroplast inheritance biparental. In such plants, defective chloroplasts are a cause of variegation: a mixture of normal and defective chloroplasts in a zygote may sort out by mitotic segregation during plant growth and development, thereby producing alternating green and white patches in leaves. The green patches contain normal chloroplasts, while the white patches contain defective chloroplasts.
Perhaps the organelle genetic systems are an evolutionary dead-end. In terms of the endosymbiont hypothesis, this would mean that the process whereby the endosymbionts transferred most of their genes to the nucleus stopped before it was complete. Further transfers may have been ruled out, for mitochondria, by recent alterations in the mitochondrial genetic code that made the remaining mitochondrial genes nonfunctional if they were transferred to the nucleus.
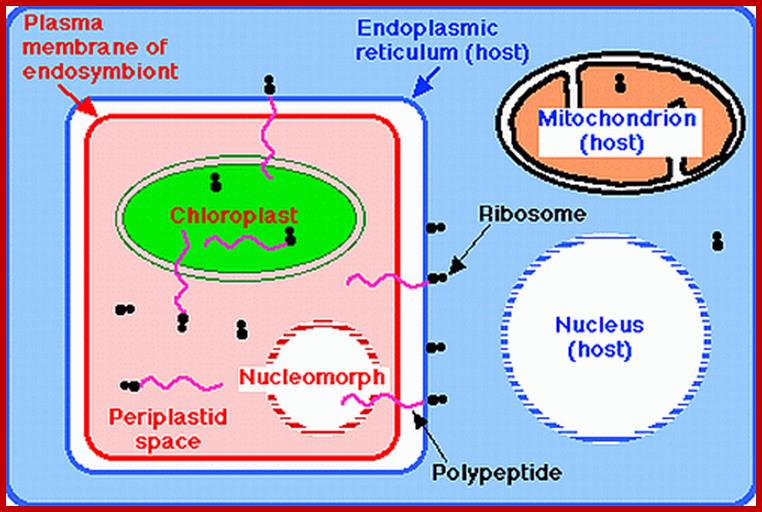
Symbiotic ode of origin of plastids; www.users.rcn.com;
For more refer. www.grkraj.org