Cell Division 1
The mechanism of cell division; Amitosis, Mitosis and Meiosis
And Cell Cycle regulation
CELL DIVISION;
Cells of all organisms undergo cell division at one or the other stages of their development. In many unicellular forms, cell division is an important mode of multiplication or calls it as reproduction. But in multi-cellular organisms, cell division is absolutely required for growth. Reproductive elements like gametes are the other important products of cell divisions.
Types of Cell divisions: Organisms exhibit two types of cell divisions. This is based on the pattern of distribution of parental chromosomes to the daughter cells. They are Mitosis and Meiosis; however, in prokaryotic organisms like bacteria and blue green algae, where there is no organized chromosomes and the nucleus; the cell division is equational and it is called Amitosis, for the mitotic apparatus and such complicated chromosomal movements are absent, but their parental DNA is separated in equal numbers. However, the genetic materials like in DNA (ssDNA or dsDNA) or genetic RNA in viruses do undergo replication cum separation, such division and separation exist in many viruses either in infected bacteria or in eukaryotic cells. Viral DNA or RNA are replicated in host cells. In most of the viruses replicated genetic material. While replication is going on they produce their respective proteins only one DNA or RNA is loaded into their newly formed capsids. Whatever may be the types, all cellular prokaryotic and eukaryotic cell divisions involve two important events viz, DNA replication, nuclear division called Karyokinesis and then cytoplasmic division called Cytokinesis.
Amitosis:
Amitosis in Eukaryotic cells- Once called closed Mitosis:
v
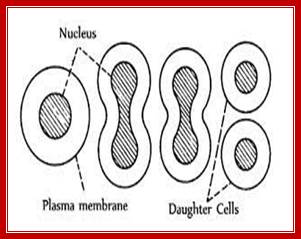
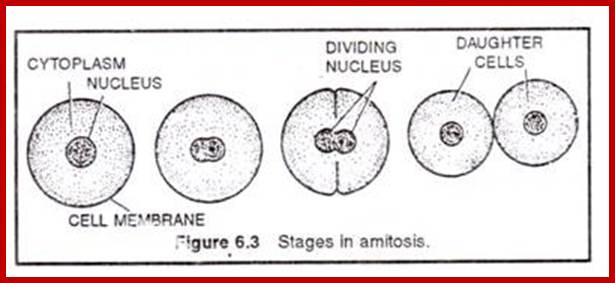
Cell division by simple cleavage of the nucleus and division of the cytoplasm without spindle formation or appearance of chromosomes. It is also called direct cell division.
Amitosis in Eukaryotic cells- Once called closed Mitosis: Why is amitosis called direct cell division? – Quora
Amitosis
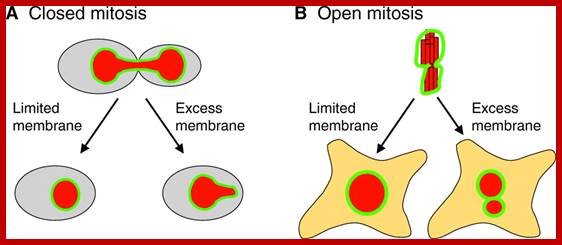
Closed and Open mitosis:
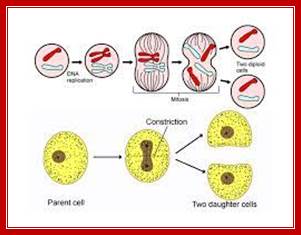
In closed mitosis, the nuclear envelope remains intact and chromosomes migrate to opposite poles of a spindle within the nucleus and the nuclear membrane cleaves in the middle and two nuclei are produced. In open mitosis, the nuclear envelope breaks down and then re-forms around homologous chromosomes separate.
Amitosis occurs in mega-nucleus of paramecium, nuclei of internodal cells of Cham, endosperm cells of seeds, cartilage cells and diseased cells. Remak (1955) discovered amitosis in RBCs of chick embryo. The term amitosis was coined by Fleming (1882).
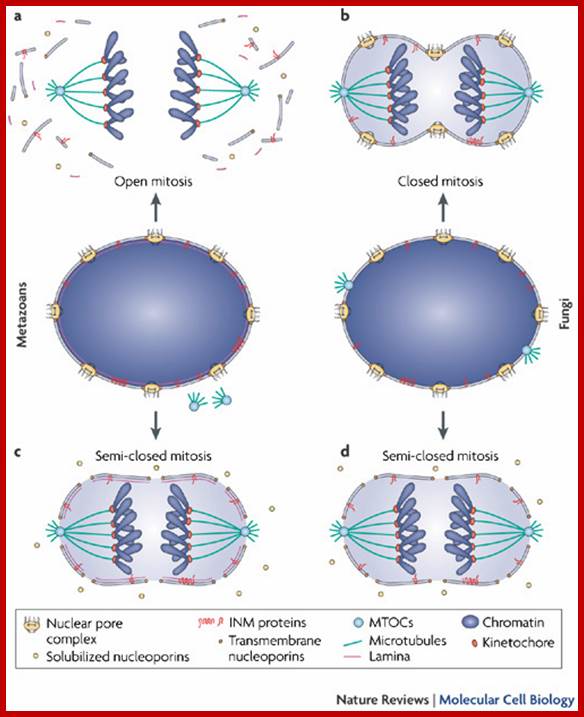
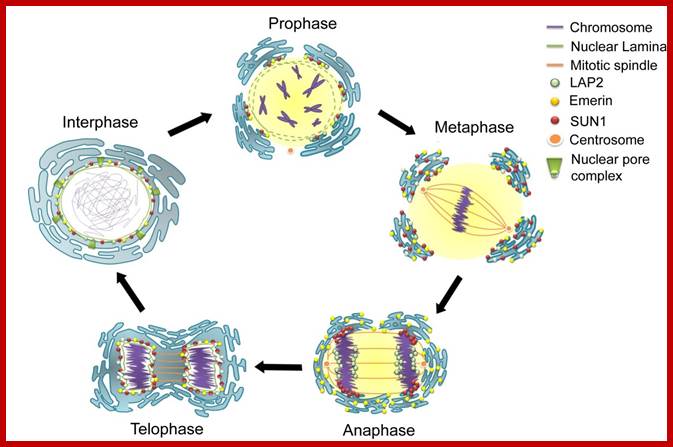
The terms 'open' and 'closed' mitosis refer to the extremes of a range of possible fates of the nuclear envelope (NE) during mitosis. a | In open mitosis, which is used in somatic cells of higher eukaryotes, the NE is completely disassembled and removed from chromatin and a cytoplasmic spindle is formed by microtubules that emanate from cytoplasmic centrosomes. b | In closed mitosis, the NE stays intact. Here, microtubule-organizing centers (MTOCs) are either constantly part of the NE (for example, in Saccharomyces cerevisiae) or are inserted into the NE during mitotic entry (for example, in Schizosaccharomyces pombe), and in both cases MTOCs direct the formation of a nuclear spindle. The establishment of a nuclear spindle requires nuclear uptake of tubulin. Closed mitosis is the most common mechanism in lower eukaryotes. A prevalent intermediate between open and closed mitosis is the partial disassembly of the NE. c | In higher eukaryotes, semi-closed mitosis is accomplished by certain cell types, such as in Caenorhabditis elegans early embryos or during syncytial embryonic divisions in Drosophila melanogaster. Here, the NE only partially opens up near to centrosomes to allow cytoplasmic spindle microtubules to reach the nuclear interior without the need for major rearrangements of NE components. In syncytial cells, this ensures that spindle microtubules capture the correct chromosomes in the common cytoplasm. The NE finally breaks down during anaphase. d | Some lower eukaryotes, such as the filamentous fungus Aspergillus nidulans, also undergo semi-closed mitosis and partially disassemble their nuclear pore complexes to achieve the rapid influx of tubulin. INM: inner nuclear membrane. Stephan Güttinger, Eva Laurell & Ulrike Kutay; http://www.nature.com/
Cell division varies in different systems. One called endocytosis, where chromosomes duplicate without dissolution of nuclear membrane, resulting in Polyploidy genome; this is called endocytosis. In another type chromosomes duplicate without separation and without nuclear membrane dissolution; this results polytene chromosomes. This process is called Endocycling. These types are found variety of organisms. Ex. Arthropods.
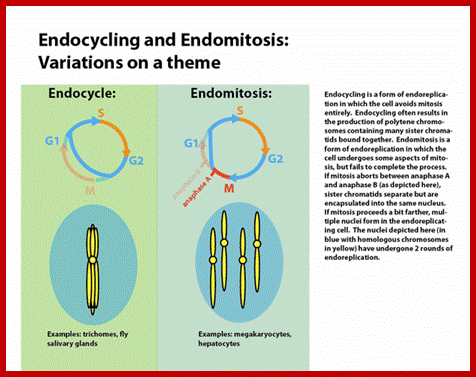
Endocycling is endo-replication without mitosis; endocycling results in the production of polytene chromosomes, https://en.wikipedia.org/
http://www.biologydiscussion.com/
The limited flat membrane hypothesis: During closed mitosis (A), excess membrane in the form of sheets results in a failure to reform a spherical nucleus, suggesting that limited membrane availability drives nuclear shape change at the end of mitosis. During open mitosis (B), excess flat membrane might facilitate the formation of multiple nuclei that collectively have the same volume as a single nucleus that would form under conditions of limited flat membrane availability. The NE is shown in green and the DNA in red. See text for more details.
The Nucleus during Mitosis: The long and viscous road: uncovering nuclear
diffusion barriers in closed mitosis: Eder Zavala and Tatiana
T. Marquez-; ttp://www.ncbi.nlm.nih.gov/
Yeast cells retain their nuclear membranes during cell division, in a process called closed mitosis. Membrane-bound proteins segregate asymmetrically in the process, with some getting localized in the mother cell and others in the bud (dots in the figure). These authors explored mechanisms by which yeast cells might prevent protein diffusion across the division plane, and hence maintain the localization. They found that a combination of protein rings and sphingolipid domains is necessary during early anaphase, but that sphingolipid domains alone are adequate during late anaphase (figure), due to the elongated nuclear neck.
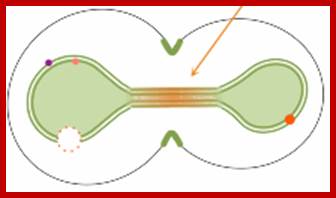
The Nucleus during Mitosis; http://www.ncbi.nlm.nih.gov/;http://www.smoldyn.org/
http://jcs.biologists.org/
Open and closed mitosis. (A) Open mitosis is so named because of the disassembly of the NE (green) during mitosis, which opens up the nucleus and exposes the chromosomes (red) to the cytoplasm. The NE breaks down early in mitosis, as the chromosomes condense, allowing microtubules (purple filaments) that emanate from centrosomes (purple structures) to associate with the chromosomes. During mitosis, the chromosomes congress to the metaphase plate, followed by separation of sister chromatids in anaphase. The NE begins to reassemble shortly thereafter, in telophase. Once the NE is completely assembled, the nucleus expands and the chromosomes return to their decondensed state in interphase. (B) Closed mitosis is so named because of the persistence of the NE throughout the cell cycle, such that the nucleus never opens to the cytoplasm. This type of mitosis occurs in certain fungi (such as budding yeast, shown here), in which the centrosome equivalents, called the spindle-pole bodies (purple), are embedded in the NE. During closed mitosis, the spindle-pole bodies nucleate microtubules within the nucleus, but as the DNA (red) begins to segregate, the nucleus has to elongate. Once segregation is completed, the nucleus divides and re-establishes a spherical shape. Note, that in budding yeast, chromosome condensation and a metaphase plate are not visible by microscopy.
Bacterial Cell division:
E. coli cells; www.unc.edu
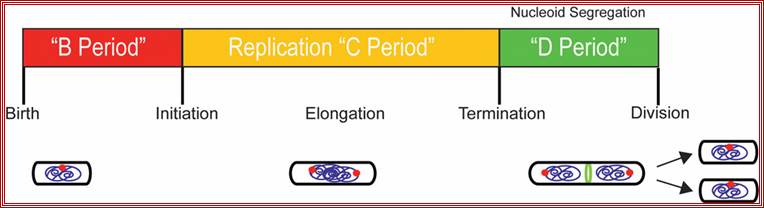
Replication takes place in three stages- Initiation, elongation and termination. Note- The daughter DNAs are attached to mesosomal membranes; cytoplasmic division leads to separation of DNAs by cleaving the cytoplasm by the Fts (complex) ring. (Amazing).
In bacteria, the DNA replication cycle (or C-period) is divided into three stages: Initiation, Elongation, and Termination. Both E. coli and B. subtilis possess an ~4 Mbp circular genome with a single origin of replication (oriC). In both organisms, C-period length is relatively constant under conditions supporting rapid growth rates (~40 minutes in E. coli cells with mass-doubling times under 60 minutes).
Replication is initiated by the highly conserved AAA+ ATPase DnaA, which binds adjacent to oriC and induces strand separation. Melting of the origin region permits the binding of DNA polymerase III (Pol III) and its accessory proteins. During elongation the replication fork proceeds bi-directionally around the chromosome, eventually reaching the terminus (terC), where the replication complex disengages from the DNA through the action of specific termination proteins.
Arrows indicate the events. https://www.ncbi.nlm.nih.gov/
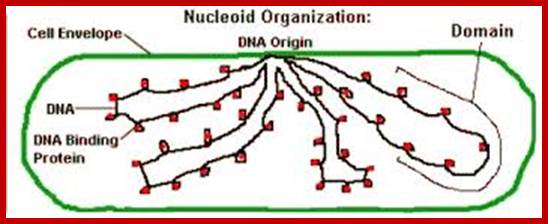
As the cell grows in size, the circular DNA molecule that is attached to the mesosome membrane; starts replicating at the initiating point called Origin OriC, which is located very near to the mesosomal attachment loci. The length of DNA is many times longer than the length of the bacterial cells; if the bacterial cell is 2micron and the DNA length can be 1.0 to 1.3 mm but entangled. DNA once replicated remains entangled, and this has to be detangled to separate them into single and independent molecules; this is achieved by Topoisomerases. Two micron sized bacterial cell contains crowded RNAs, proteins and metabolites filled in its cytoplasm; in such molecular density the duplicated DNA has to be separated and moved to daughter cells. Bacterial DNA is coiled to each other in tight bundle like a ‘rosette’, from which some strands project out with RNA strands still bound to DNA.
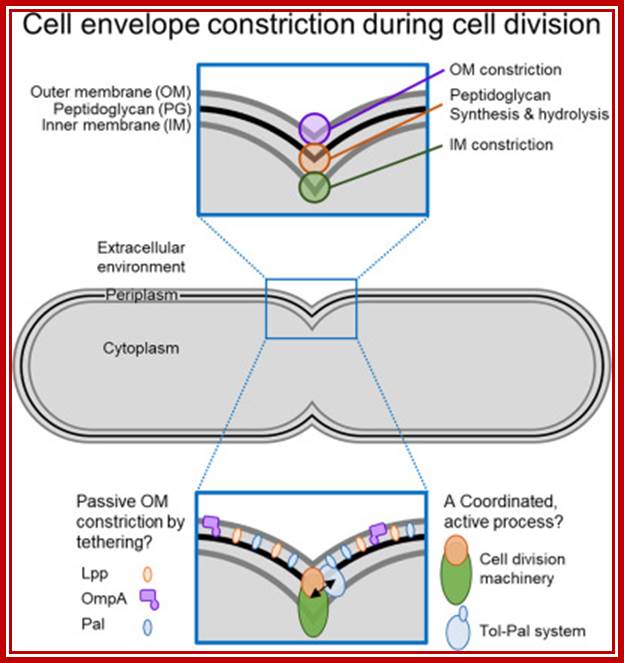
Bacterial cell produces a constriction and daughter DNA molecules separate, they are still attached to membranes and the cells separate. Constriction leads to the separation of cells.
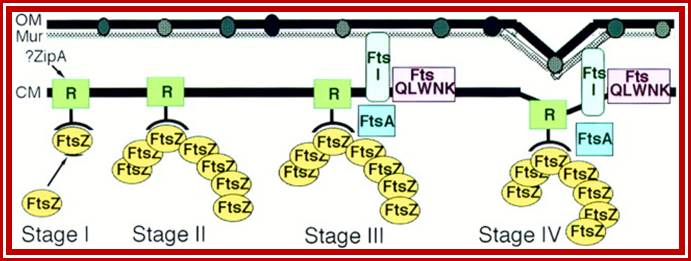
Note- FtsZ is a complex of six or more Fts-proteins.
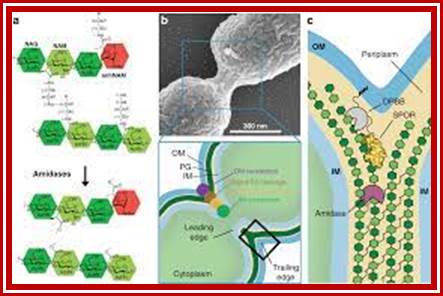
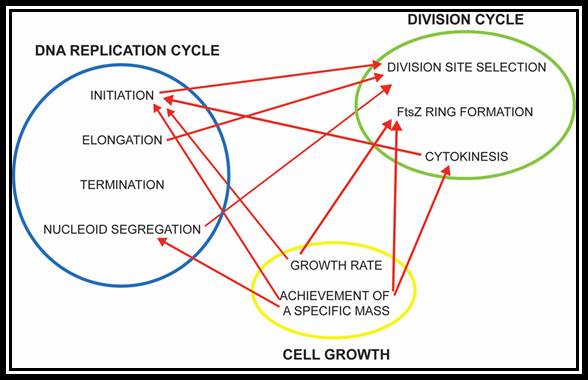
The bacterial cell cycle can be arbitrarily divided into two segments: a DNA cycle that includes DNA replication and chromosome segregation, and a division cycle that leads to cytokinesis and cell separation. During the division cycle, the cell must identify the mid-cell site at which division later occurs, differentiate this site in preparation for cytokinesis, and finally form the division septum by the coordinate ingrowth of the cytoplasmic membrane, the rigid murein (peptidoglycan) layer, and, in Gram-negative bacteria such as Escherichia coli, the outer membrane of the cell envelope. Recent advances have led to an increased understanding of important elements of this complex series of events.

Microstructures in the bacterial cell. Structures such as the chemoreceptor array, the FtsZ ring (complex of 6-7 Fts proteins), and the flagellar motor are made of multiple subunits operating in a coordinated fashion. Many of these assembled structures have been observed directly in electron microscopy images (23, 61, 95, 96). The properties of these structures are unique because the mechanics and chemistry in the subunits are coupled. Subunits are also mechanically coupled to each other, leading to cooperative effects. The bacterium is not just a microbe?! https://www.researchgate.net/, viruses replicating within the bacterium is the most, and most amazing.
Many, although probably not all, of the proteins involved in the division cycle of E. coli are now known. One of these proteins, FtsZ, is now recognized to play a key role in the assembly of the division apparatus and in the process of cytokinesis. It’s wide distribution and high degree of sequence conservation suggest that it probably plays a similar role in all bacterial and archaeal species. In this minireview, we discuss the likely sequence of events that occurs during differentiation of the division apparatus of E. coli, beginning with the localization of FtsZ at the potential division site and ending with the generation of two new daughter cells.
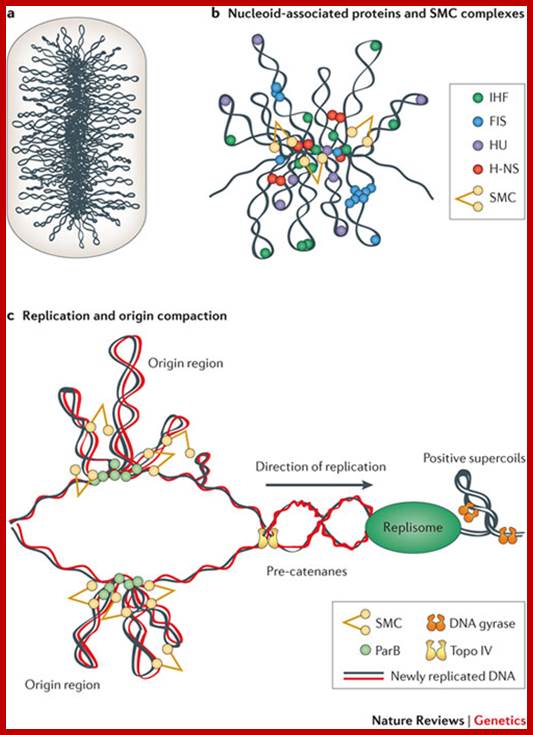
Bacterial DNA as Nucleoid; circular DNA folded and Plasmids; Xindan Wang, Paula Montero Llopis & David Z. Rudner ;www.biotechlearn.org.n
Bacterial supercoiled DNA is compressed into bottle brush mode with a dense core from which the loops many of 10kb size loop out; http://www.nature.com/

The “rosette” model of DNA organization. Electron micrograph of isolated membrane-free chromosomes of E. coli. The central core, from which several 100 or more independent loops radiate, is sensitive to RNAse; http://www.ncbi.nlm.nih.gov/
Separation of highly condensed DNA is facilitated by the binding of ParB protein (in Caulobacter) to a site near origin (ori C) which is localized to cell poles. It is an amazing process.

Chromosome organization in a model bacterium; (A) The Caulobacter chromosome is linearly organized, and anchored to the flagellated pole viapar S/ParB/Pop Z. (B) In Vibrio cholerae, the origin region of the larger chromosome (chromosome I) is localized to the cell pole, whereas the origin of the smaller chromosome is localized to the cell center. The organization of the bulk of the chromosomes, as well as their separation or intermingling, are currently not known. (C) Four loci have been localized in vegetative cells of Bacillus subtilis, and their organization is reminiscent of the linear order seen in Caulobacter. Although the origin region is localized near to one pole, it appears not to be anchored to the cell membrane. (D) Sporulating cells of B. subtilis, however, do anchor the origin region, through RacA/DivIVA, to the negatively curved membrane at the pole. RacA also binds all along the chromosome, compacting it into a long “axial filament” before sporulation. (E) The E. coli origin localizes to mid-cell, and the two replichores are separated into opposite cell halves. The terminus is broadly localized (arrows), and may be found on either side of the cell center. http://www.ncbi.nlm.nih.gov/
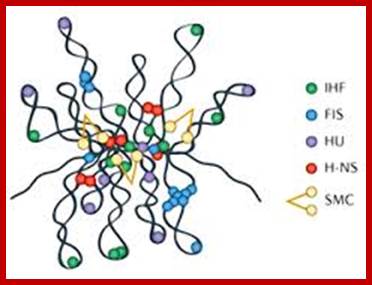
Nucleoids; www.schaechter.asmblog.org
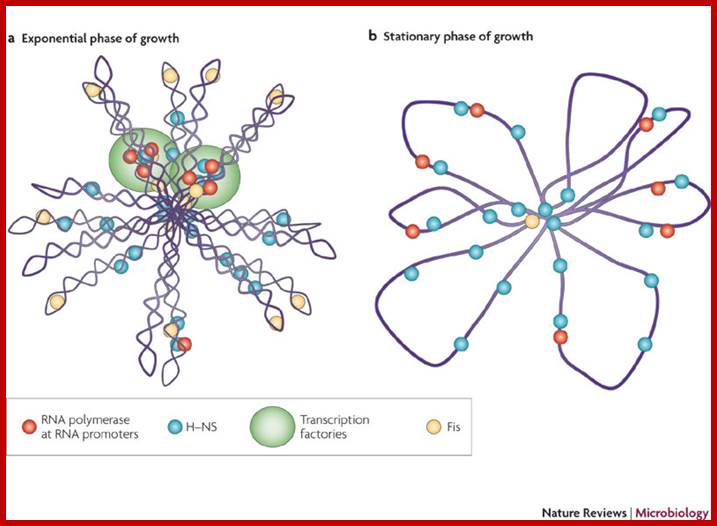
The folded bacterial DNA loops or nucleoid DNA is associated with their H-Ns histone like proteins (15.6kDa) which act like pleotropic regulator of gene expression; The H-Ns has found to act at least 250 DNA loci covering >more than 1000 genes. It acts like repressor. Bacterial Histone like another protein HU 10kDa acts similar to eukaryotic H2B, by binding to DNA it induces negative supercoils in bacterial DNA; http://www.nature.com/
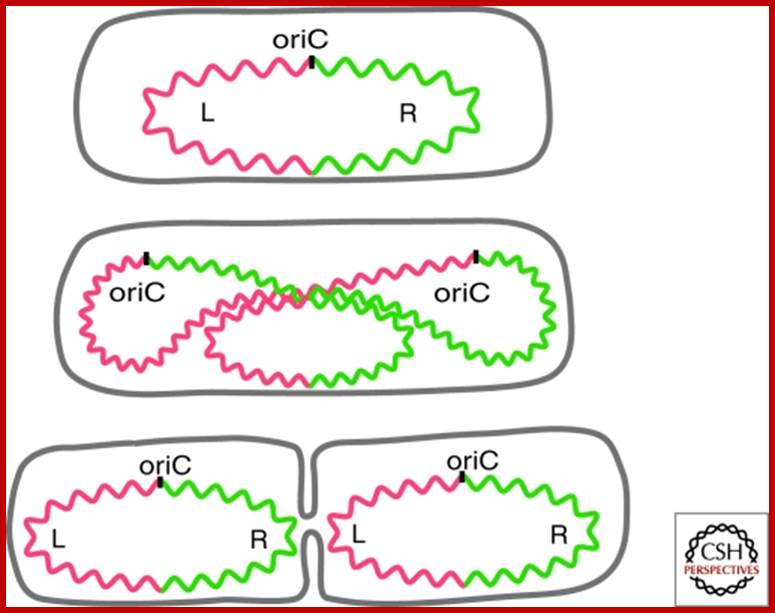
Chromosome organization in E. coli; The origin of replication of the E. coli chromosome (oriC) is located at mid cell, and each arm is kept in a separate cell half (top). The terminus region is broadly distributed along the long axis of the cell (not shown). As replication proceeds (middle), each daughter oriC is segregated to the cell quarters and, when replication is complete, the daughter chromosomes adopt a transnationally symmetric <L-R-L-R> configuration (bottom).
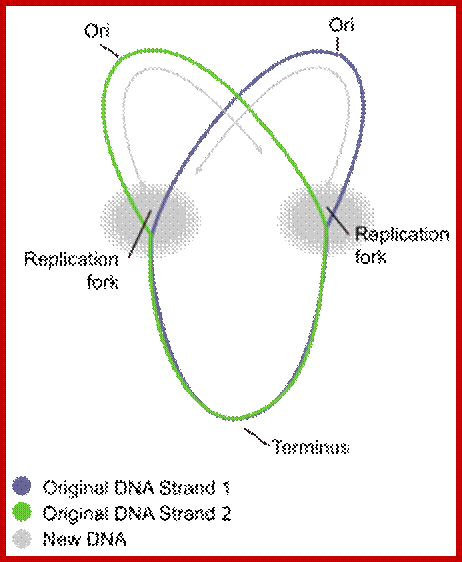
Circular bacterial DNA replication starts at origin and proceeds in bidirectional manner; each half of the chromosome replicated called ‘replichore’ ps; http://en.wikipedia.org
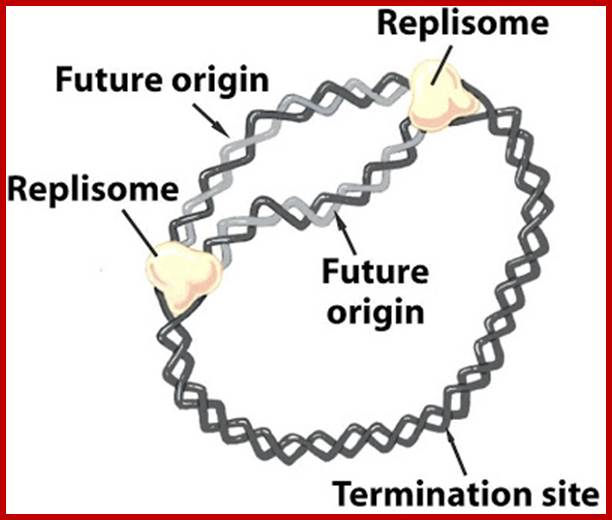
The replisome assembles at origin and then moves bidirectionally until they meet at terminal region http://sandwalk.blogspot.in/
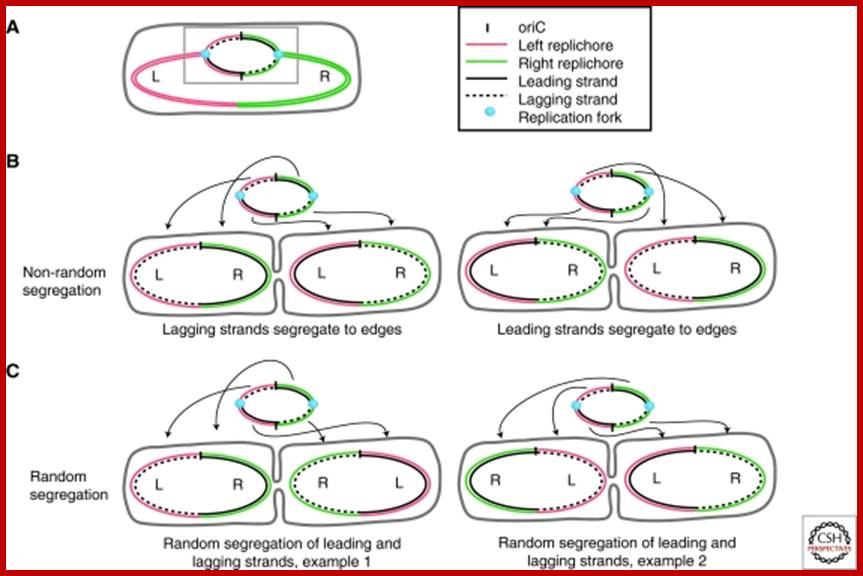
Non-random segregation in E. coli: (A) A replicating mother cell is shown, highlighting the Left and Right replichores as well as the difference between leading-strand-replicated (solid black lines) and lagging-strand-replicated DNA (dashed black lines). The replication bubble (grey box) is also shown, and repeated above each example in B and C to illustrate the origin of each configuration. (B) To achieve the <L-R-L-R> configuration seen, it is necessary for both lagging strands and both leading strands to be segregated to the distal edges of the cell. Note that in ∼90% of cases, leading strand segregation to the distal edges (right) is observed. (C) If random segregation of leading and lagging strands is imposed, the <L-R-L-R> configuration cannot be achieved, and mirror symmetry appears (e.g., <L-R-R-L>). http://www.ncbi.nlm.nih.gov/
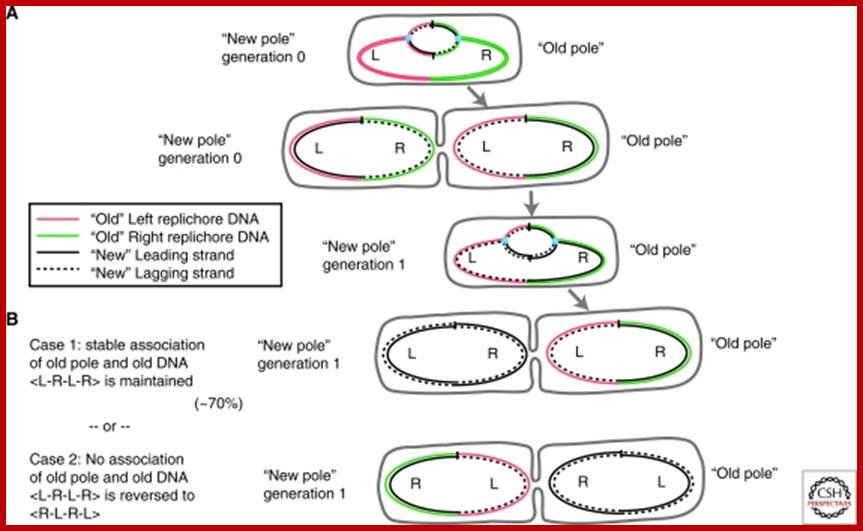
“Immortal strand” inheritance versus Leading strand segregation. Two generations of DNA replication and segregation are shown, to illustrate the association of an old DNA strand (coloured) with the old cell pole. Note that in all cases, leading strand segregation to the distal cell edges is maintained. (A) After the first generation, both daughter cells carry one “new” and one “old” strand of DNA (black and coloured, respectively). (B) During the second round of segregation, two scenarios are possible: (1) immortal strand segregation is kept, and the “old” pole stays associated with the “old” (coloured) strand of DNA (top). Note that this is the case observed for E. coli cells in ∼70% of cases; (2) Immortal strand segregation is not kept, and the “old” pole received two “new” strands of DNA. In this case, the configuration of the chromosome changes from <L-R-L-R> to <R-L-R-L>. http://www.ncbi.nlm.nih.gov/
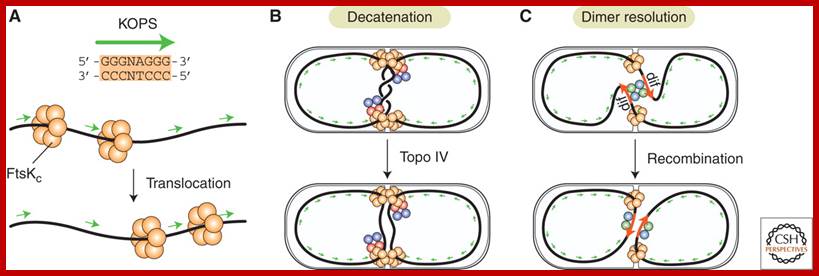
The chemical components of mesosomes are responsible for initiating replication, which will be completed in about ~15 minutes. Then the daughter molecules, still attached to the membrane, open out and segregate. A little later, almost in the middle region of the cellular cytoplasm, the plasma membrane produces an inward invagination all-round and it progresses forward till the inwardly growing membranes fuse with one another in the center. Proteins such as FtsZ, 40kDa (similar to eukaryotic tubulins) and actin- related proteins called FtsA are involved in the formation of Z-ring in the middle of the cell. It also requires proteins such as ParB, Par A and ParC (sop ABC). ParB binds near Ori-C and it is bipolarly localized for partitioning DNA to poles. This results in the partition of cytoplasm into two compartments. Soon, the newly formed plasma lemma loaded with components secretes the cell wall materials into the space found between them. Then the middle wall splits across in the middle and two daughter cells separate.
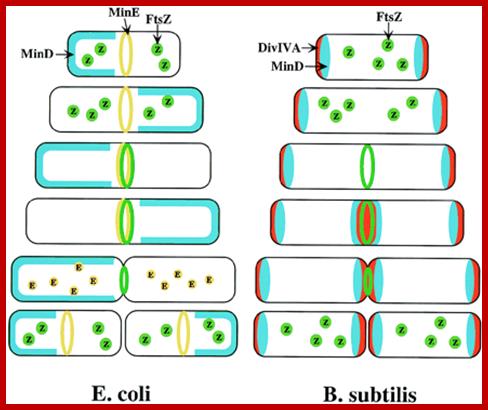
Models for division-site selection in E. coli (Left) and B. subtilis (Right). MinD is in blue, MinE in yellow, FtsZ in green and Div-IV in red. Shown are different stages of the cell cycle, beginning with a newborn cell and finishing with cell division that produces two daughter cells. (Left) In E. coli, MinE localizes to a ring-like structure at or near the middle of the cell early in the division cycle. MinD accumulates alternately at the membrane periphery on either side of the MinE ring (3). The alternation of MinD localization from one pole to the other occurs at a frequency of the order of tens of seconds. The rapid relocation of MinD ensures that no FtsZ ring is assembled at either the ¼ or ¾ sites in the cell halves. The presence of MinE at midcell prevents the MinD inhibitory activity at this site, allowing assembly of the FtsZ ring at this site. The MinE ring disassembles before completion of constriction. (Right) In B. subtilis, DivIVA and MinD are localized to the cell poles in a newborn cell, and therefore the presence of the MinD inhibitor prevents the formation of the FtsZ ring at these sites. Later, presumably after completion of DNA replication, a new potential division site is created at midcell. The sequestration of the MinD inhibitor to the poles allows assembly of the FtsZ ring at mid-cell and recruitment of other cell division proteins. At this point, the division machinery presumably becomes resistant to the MinD inhibition. DivIVA and MinD proteins then are recruited to the mid-cell. Constriction then is initiated. When constriction is completed, the FtsZ ring disassembles, but DivIVA and MinD remain at the newly formed poles, preventing further divisions from taking place in these polar sites. http://www.pnas.org/
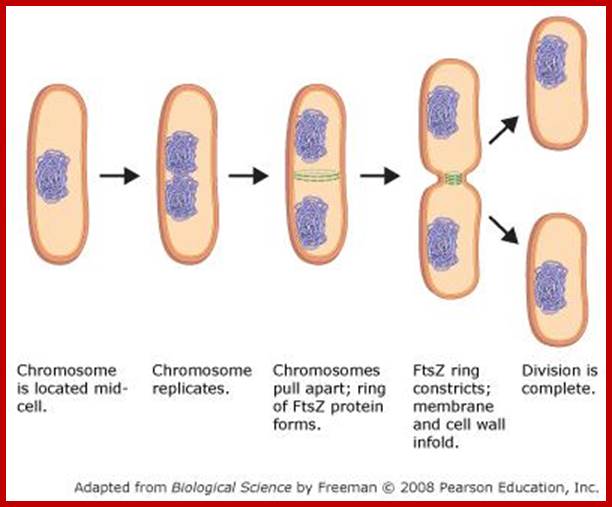
It is important to note that this type of cell division does not involve any complicated structural movements as found in eukaryote cells but the cell undergoes DNA duplication and cytoplasmic division by cleavage.
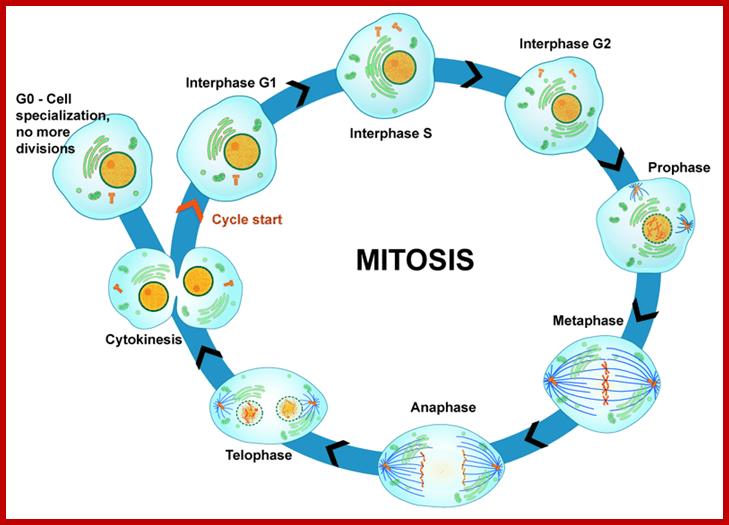
MITOSIS-Eukaryotic Cells
Multicellular organisms start their development as a unicellular zygote. Under favourable conditions unicellular organisms multiply and produce a huge population. A fertilized egg in some animals may develop into a giant plant or animals such as an elephant or sea animal. Some haploid organisms produce spores or gametes as a means of reproduction. All the above processes are achieved by cell divisions. This process involves equal distribution of genetic material. But in meiosis the genetic material is reduced to half of the original in the first division of meiosis.
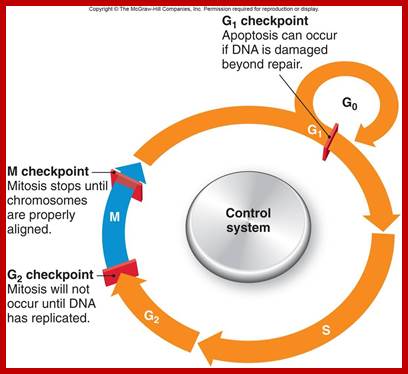
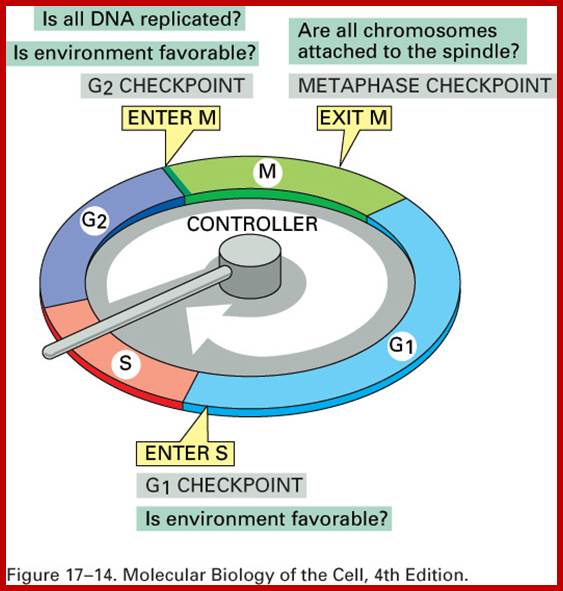
Eukaryotic cell division is regulated by check point system; https://www.studyblue.com

Plant cell division; www.csus.edu
Animal cell division; http://www.ck12.org/
Cell cycle control proteins monitor and act at specific stages of cell cycle; http://www.pha.jhu.edu/
The process involves two important steps. The first is the division of nucleus (karyokinesis) and the second which normally follows is called cytokinesis. Cytokinesis depending upon the cell type (organism) and stage at which it produces two equal cells or cells show asymmetric or polarized cell division. In some species only nuclei divide they are called coenocytes ex. Few fungi.
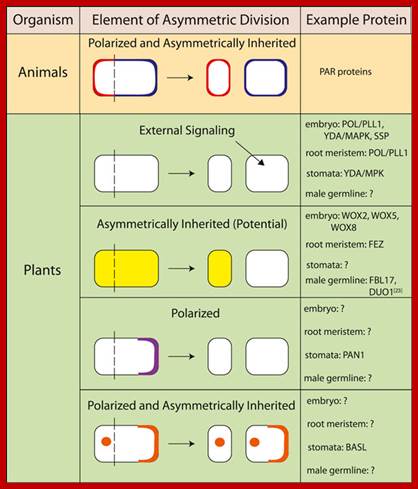
Animal and plant proteins that demonstrate various elements of asymmetric division; BASL, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE; DUO1, duo pollen 1; FBL17, F-box like protein 17; MAPK, mitogen-activated protein kinase; MPK, Arabidopsis gene encoding a mitogen-activated protein kinase; PAN1, PANGLOSS 1; PAR, partitioning defective; PLL1, POLTERGEIST-LIKE 1; POL, POLTERGEIST; SSP, SHORT SUSPENSOR; WOX, WUSCHEL-related homeobox; YDA, YODA; Asymmetric cell division; http://f1000.com/
Furthermore, depending upon the presence or absence of astral elements, it has been classified into astral type and anastral type. However, the whole process of cell cycle progresses sequentially through different stages, which merge with one another smoothly. Basing on the complex biochemical, molecular and physical changes, different stages have been recognized. The stages are interphase-Gᴼ, G1, S and G2, M-phase-(prophase, metaphase, anaphase, and telophase) and finally cytokinesis. In many cytokinesis does not take place and generates coenocytic cells.
The time required for these different stages vary from cell type to cell type and organisms to organisms. Nevertheless, the interphase is the longest phase and the most variable. For example: in the root meristems of Vicia faba and Pisum sativum, the total time required for the whole cell cycle is about 24 hours, out of which interphase occupies about 21 hours and a half. On the other hand prophase to telophase requires just about two hours and a half. On the contrary, in the corneal epithelial cells of rats, the whole process requires just 60 minutes whereas the interphase takes up 14-24 hours.
Interphase: In yester years, interphase was considered as the resting stage, but actual resting stage is quiescent stage and it is Go but when the cell is stimulated and preparing for cell division, the cell become very active and the cell prepares itself for the entire proceedings of cell division. Because of its long duration and varied biochemical activities, this stage has been further sub-divided into phases like G1, S, G2 and M of which G1 phase is the most variable in duration. Intense biochemical activities take places at this stage and all precursors for DNA synthesis, histone synthesis, assembly of proteins, energy rich molecules required and many others are mobilized from cytoplasm. However, as cell is stimulated for cell division cell enters the G1 stage and the cell division is on; single stranded chromosomes move on to the next stage called ‘S’ phase. At this stage chromosome undergo unwinding and its chromosomal DNA undergoes replication and chromosomal components reassemble. During this process, the long chromatin DNA double helix unwinds and semi-conservative replication is initiated at several points simultaneously. Entry into each of the stages and exit from each of the phases is regulated and monitored by what is called Check point proteins.
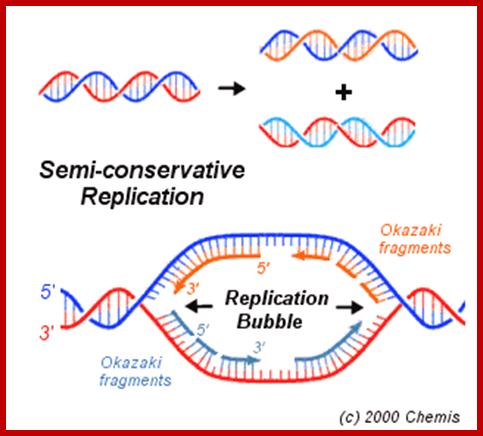
www.rrresearch.fildofscience.com

https://www.studyblue.com
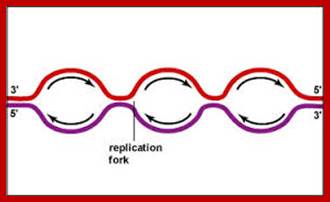
Then the replication fork progresses in both directions (bidirectional) till they meet the neighbouring replicons. During replication, one of the daughter DNA molecules retains the parental histone core proteins and the other gets associated with the newly synthesized histone units imported into the nucleus to form a new chromatin thread. Thus two Chromatin strands are formed in about seven hours of time, while G1 stage takes about 5 hours. Once the chromatin threads are duplicated, G2 phase is initiated. At this stage intense biochemical activities required for chromosomal contraction and development of mitotic apparatus. This state lasts for about 3 hours. All these activities ultimately result in the increase of nuclear size and now the cell is set to enter into next phase. It is important to note that throughout this stage various types of RNAs are synthesized. Even as the chromatin DNA is undergoing replication, RNA synthesis and protein synthesis continues.
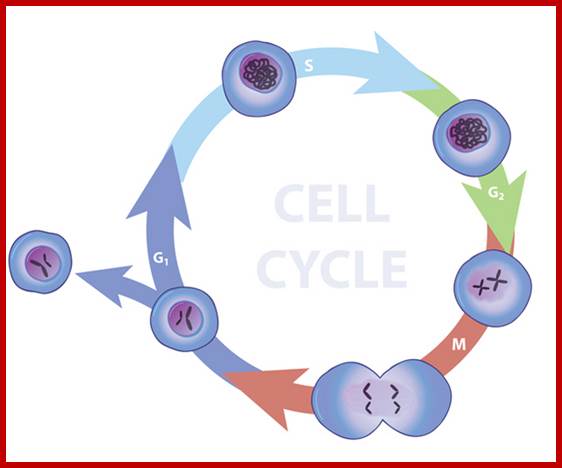
Nature Education; http://www.nature.com/scitable
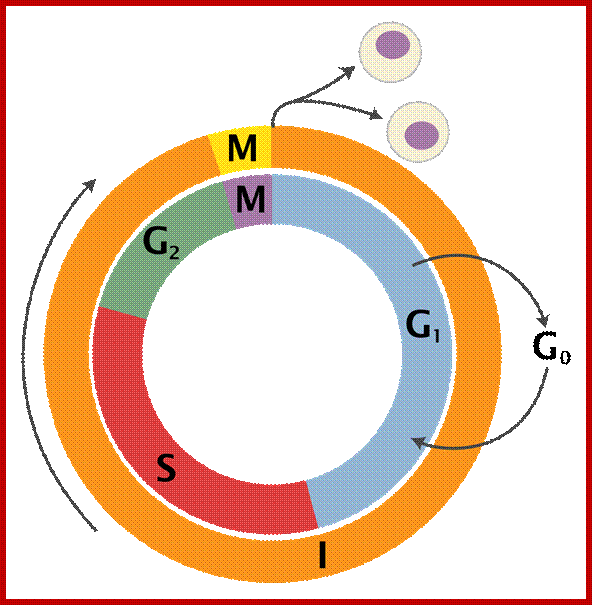
www.en.wiikipedia.org
Schematic of the cell cycle; outer ring: I=Interphase, M=Mitosis; inner ring: M=Mitosis; G1=Gap phase 1; S=Synthesis; G2=Gap phase 2. The duration of mitosis in relation to the other phases has been exaggerated in this diagram.
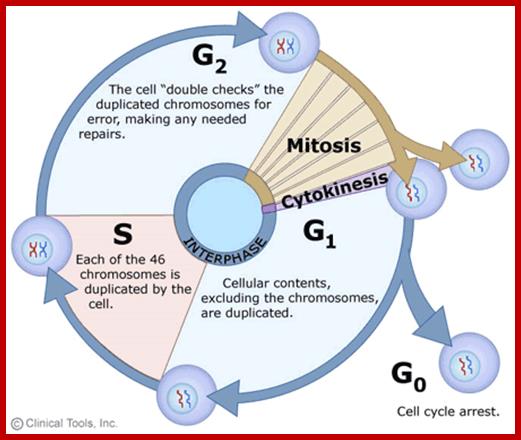
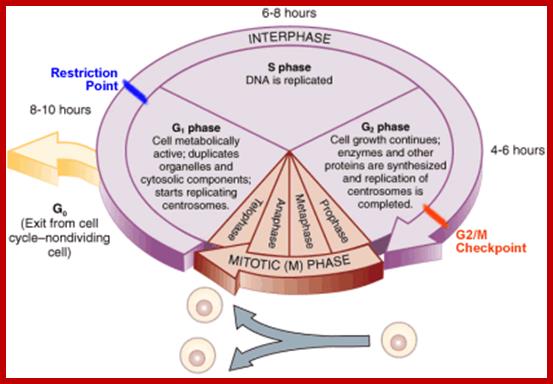
Cell cycle duration of each stage; Go 8-10hrs, S-phase 6-8hrs, G2 phase 4-6hrs, M-phase 2-4hrs; entry into cell cycle atG1 phase it is regulated by RESTRICTION POINT, Check point at G2/M
www2.le.ac.uk;www2.le.ac.uk; http://www.angelfire.com/
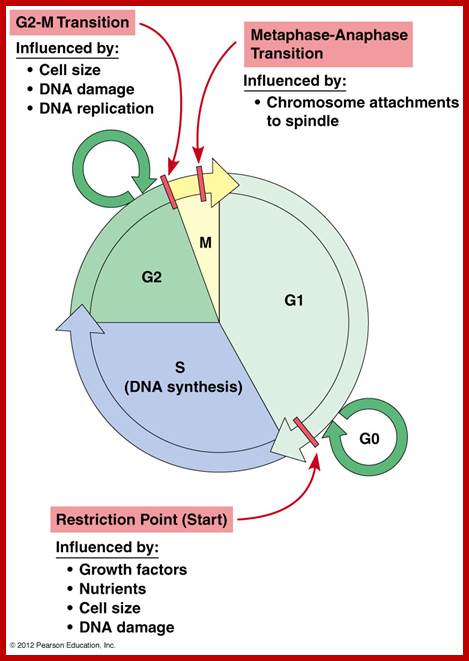
http://www.mun.ca/biology
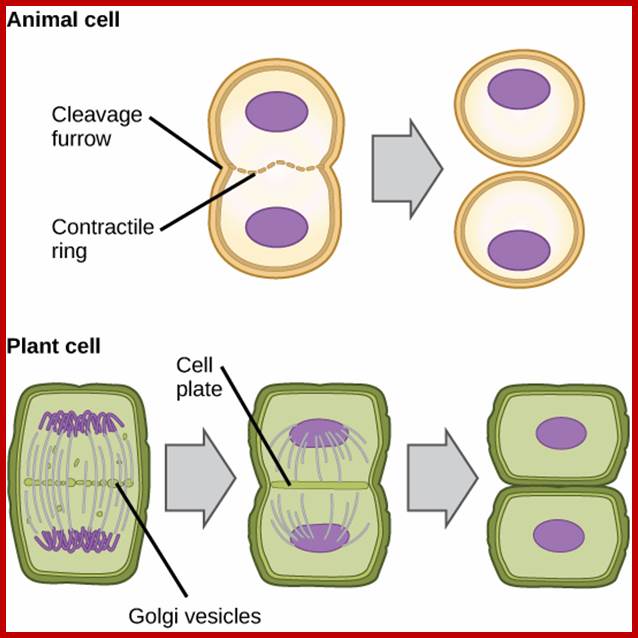
Differences between Animal and plant cell division-cytokinesis; https://www.boundless.com
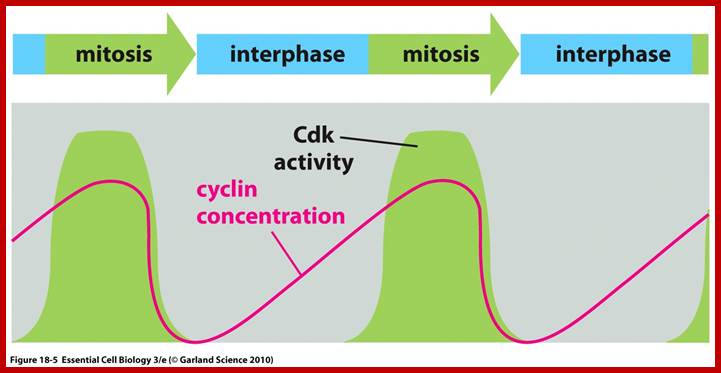
Cell division requires sever proteins as factors for initiation and progression. The most important factors are cyclins and CDKs (Cyclin Dependent Kinase). Cyclins of different types bind to specific CDKs (shown in the figure below).
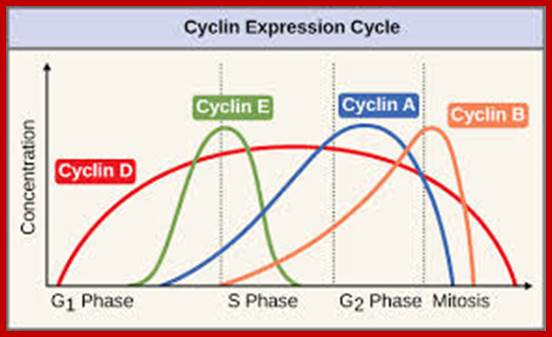
Regulator molecules of the cell cycle; www.boundless.com; www.philschatz.com
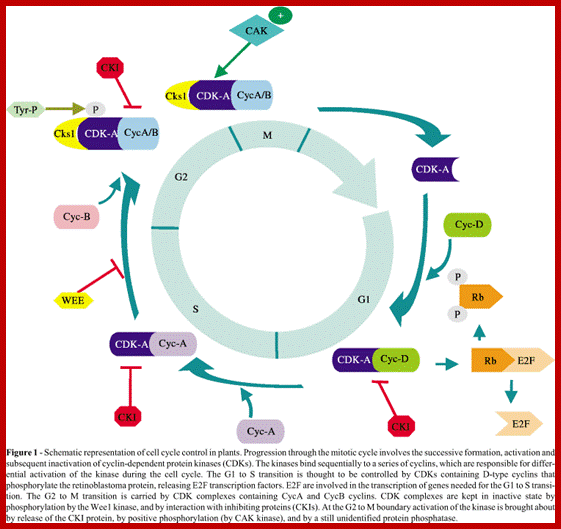
Cell cycle has check points at specific phases, until the check points cleared, the cell remains at the said stage. Action of different Cyclins and specific CDKs (Kinases) at specific phases and they actually regulate the steps; check point protein monitors the correctness of each phase;
www.oregonstate.edu; and www.scielo.br
Cyclin Protein-general; https://en.wikipedia.org/wiki/
CDK4-protein-general; https://en.wikipedia.org/wiki/

Diagrmatic view of Cyclin-A and Cdk2-proteins in ribbon model; Red binding sites for AMP/PNP; https://en.wikipedia.org/wiki/
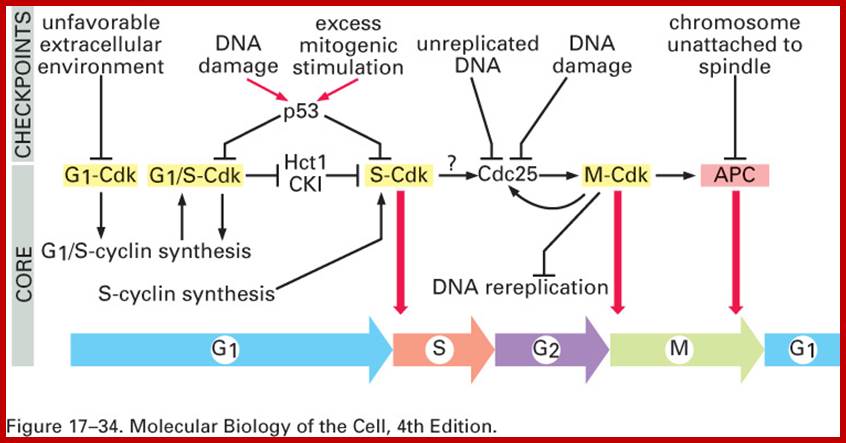
Note at what stage and what check points are operated; Phase or stage specific cyclins and CDKs operate at specific stages; G1 cyclins and CDKs, S-cyclins and CDKs M-cdc25 and M-cCdks; The most important protein that operates during cell cycle is P53 which operates when DNA is damaged and APC operates when chromosomes remain not attached to centromeres; Their activity is also regulated by CDK inhibitors called CKI/KIP family; www.pha.jhu.edu
Cyclin’ Main groups;
There are two main groups of cyclins:
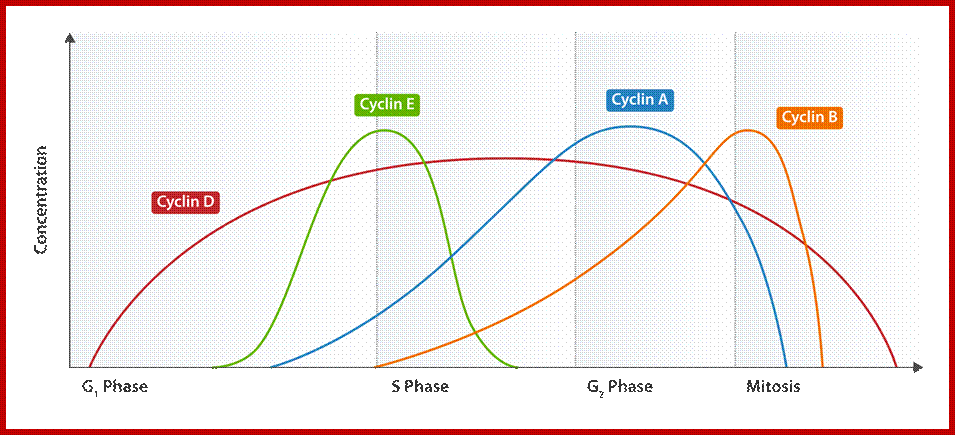
G1/S cyclins – essential for the control of the cell cycle at the G1/S transition, Cyclin D / CDK4, Cyclin D / CDK6, and Cyclin E / CDK2 – regulates transition from G1 to S phase, Cyclin A / CDK2 – active in S phase;
G2/M cyclins – essential for the control of the cell cycle at the G2/M transition (Mitosis)transition; G2/M cyclins accumulate steadily during G2 and are abruptly destroyed as cells exit from mitosis (at the end of the M-phase). Cyclin B / CDK1 – regulates progression from G2 to M phase. Subtypes cyclin include the following:
|
Species |
G1 |
G1/S |
S |
M |
|
Cln3 (Cdk1) |
Cln 1,2 (Cdk1) |
Clb 5,6 (Cdk1) |
Clb 1,2,3,4 (Cdk 1) |
|
|
Puc1? (Cdk1) |
Puc1, Cig1? (Cdk1) |
Cig2, Cig1? (Cdk1) |
Cdc13 (Cdk1) |
|
|
cyclin D (Cdk4) |
cyclin E (Cdk2) |
cyclin E, A (Cdk2,1) |
cyclin A, B, B3 (Cdk1) |
|
|
either not known or not present |
cyclin E (Cdk2) |
cyclin E, A (Cdk2,1) |
cyclin A, B, B3 (Cdk1) |
|
https://en.wikipedia.org/wiki/Cyclin
A list of CDKs with their regulator protein, cyclin or others.
· CDK4; cyclin D1, cyclin D2, cyclin D3
· CDK5; CDK5R1, CDK5R2. See also CDKL5.
· CDK6; cyclin D1, cyclin D2, cyclin D3
· CDK9; cyclin T1, cyclin T2a, cyclin T2b, cyclin K
· CDK10
· CDK13 (CDC2L5) ; cyclin L (Wikipedia)
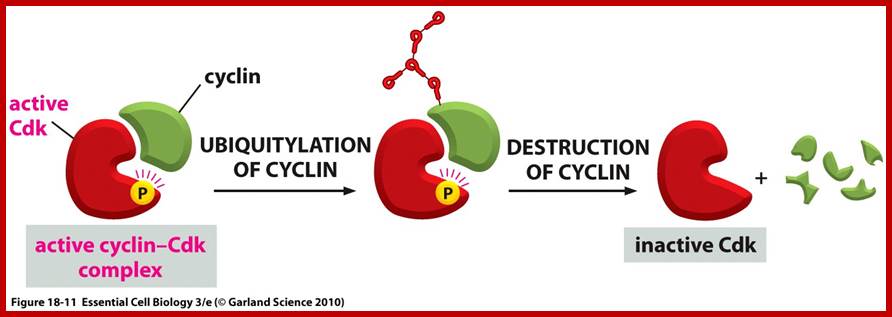
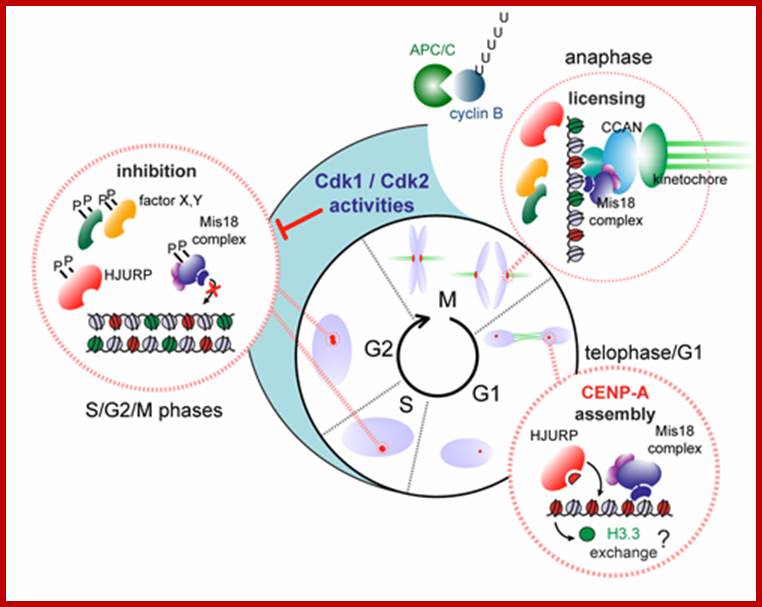
Model illustrating cell-cycle coupling of CENP-A assembly. In our 2012 Developmental Cell paper, we have uncovered the basic mechanism of G1 restricted CENP-A chromatin formation. The CENP-A assembly machinery is poised for activity throughout the cell cycle but is kept in an inactive state by inhibitory Cdk2 and Cdk1 activity during S, G2 and mitosis. Sequential loss of Cdk2 and Cdk1 activity during mitotic exit trigger CENP-A assembly. In this way the propagation of centromeres occurs only when cells have successfully duplicated their genome and executed mitosis, thereby maintaining a balance between epigenetic centromere propagation and cell division. http://sites.igc.gulbenkian.pt/
|
Species |
Name |
Original name |
Size (amino acids) |
Function |
|
Cdk1 |
Cdc28 |
298 |
All cell-cycle stages |
|
|
Cdk1 |
Cdc2 |
297 |
All cell-cycle stages |
|
|
Cdk1 |
Cdc2 |
297 |
M stage |
|
|
Cdk2 |
Cdc2c |
314 |
G1/S, S, possibly M |
|
|
Cdk4 |
Cdk4/6 |
317 |
G1, promotes growth |
|
|
Cdk1 |
Cdc2 |
301 |
M |
|
|
Cdk2 |
297 |
S, possibly M |
||
|
Cdk1 |
Cdc2 |
297 |
M |
|
|
Cdk2 |
298 |
G1, S, possibly M |
||
|
Cdk4 |
301 |
G1 |
||
|
Cdk6 |
326 |
G1 |
Orderly progression through these cell-cycle phases is controlled by the sequential activation of the Cyclin-dependent kinases (Cdks) Cdk4/6, Cdk2 and cyclin Cdc2. Their activity is regulated by various factors, including the synthesis and binding of a specific regulatory subunit (called a Cyclin), both inhibitory and activating phosphorylation events, and the association/dissociation of inhibitory molecules called Cdk inhibitors (CDIs). Mitogenic growth factors exert their effect by promoting the synthesis of the D-type cyclins and their assembly into active Cdk4/6–cyclin D complexes. By contrast, the expression of cyclin E is triggered by internal signalling pathways and the appearance of Cdk2–cyclin E kinase activity seems to be synonymous with the restriction point. The ordered activation of the remaining Cdk–cyclin complexes seem to be self-regulating: each Cdk–cyclin complexes trigger the activation of the next Cdk–cyclin species and also induces its own destruction. Under conditions of cellular stress, cell-cycle progression is disrupted by the activation of checkpoint pathways that ultimately lead to the inhibition of one or more Cdk–cyclin complexes. Jeffrey M. Trimarchi & Jacqueline A. Lees; Nature .com
Difference between Plant cytokinesis and Animal cytokinesis
|
Animal Mitosis |
Plant mitosis |
|
Centriole present |
Centriole absent |
|
Aster develops at each centriole |
Aster does not form |
|
Spindle formed-Astral type |
Spindle-non astral type |
|
Cytokinesis by furrowing or constriction |
Cytokinesis by central plate formation |
|
Mitosis occurs in all cells throughout body whenever required. |
Mitosis occurs mostly in meristems and during renewed cell division. |
|
|
|
https://en.wikipedia.org
Cdk/cyclin complexes regulate Rb/E2F- and FoxM1-mediated transcription. During the G1 phase of the cell cycle, Cdk4/cyclin D (cycD) and Cdk2/cyclin E (cycE) complexes sequentially phosphorylate (P) Rb, leading to the activation of E2F proteins and the expression of E2F-responsive genes. This cluster of genes encodes cell cycle regulators required for G1/S transition [cyclin E, cyclin A (cycA) and Cdk1], enzymes involved in nucleotide biosynthesis [thymidine kinase (TK)] and components of the DNA replication machinery [Cdc6 and origin recognition complex subunit 1 (Orc1)]. During the G2 phase of the cell cycle, Cdk2/cyclin A and Cdk1/cyclin B (cycB) complexes sequentially phosphorylate FoxM1, leading to the relief of its self-inhibition and the recruitment of a histone deacetylase p300/CREB binding protein (CBP) that activates the expression of FoxM1 target genes. This cluster of genes encodes cell cycle regulators required for the execution of mitosis (cyclin B) and interactors of the kinetochore complex crucial for proper chromosome segregation [centromere protein F (Cenpf)]. The effects of Cdk phosphorylation on FoxM1 can be counteracted by the phosphatase PP2A/B55α; http://dev.biologists.org/content.
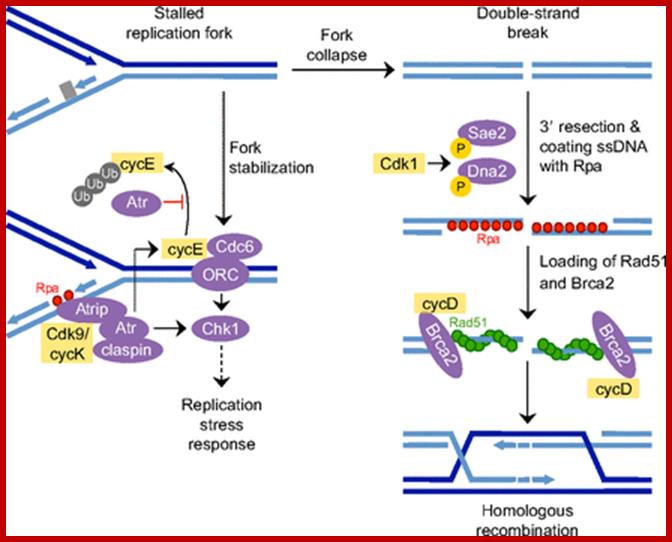
Cell cycle regulators influence DNA damage repair. In response to DNA lesions (gray box), the replication fork is stalled and the replication stress response (RSR) is initiated to prevent further cell cycle progression and replication origin firing. This is crucial for replication fork stabilization and eventual recovery from the obstruction. RSR results in the activation of Atr, which inhibits the ubiquitin (Ub)-mediated degradation of cyclin E1 (cycE). Elevated cyclin E causes the retention of Cdc6 at the pre-replication complex, which prevents the initiation of replication and activates Chk1. Through an unknown mechanism, Cdk9/cyclin K (cycK) complexes reportedly associate with Atrip, Atr and claspin to limit the amount of single-stranded DNA (ssDNA) available for replication protein A (Rpa; red circles) binding, thereby contributing to the maintenance of fork stability. In the event that the fork collapses, double-strand breaks (DSBs) are generated and these can be repaired by homologous recombination (HR). The initial step in HR is DSB resection to produce ssDNA coated with Rpa (red circles). This event is stimulated by Cdk1-dependent phosphorylation of the nucleases Sae2 and Dna2. Cyclin D1 (cycD) subsequently binds to resected DNA through Brca2 to facilitate the recruitment of the DNA recombinase Rad51 (green circles), which displaces Rpa to form the nucleoprotein filament. This marks the beginning of homology search and strand invasion during HR. ORC, origin recognition complex. http://dev.biologists.org/
Prophase:
Prophase sets in as cell cycle factors operating in regulating cell division events are ready to go. The cell volume as it reaches a critical dimension with all its components are build up, the nucleus meanwhile also enlarges as the chromosomal DNA undergoes replication and the daughter chromatids fully formed, the nuclear membrane starts dismemberment aided by lamin components as small membrane vesicles including pore complexes. Appearance of mitotic apparatus and the dismemberment of nuclear membrane coincide.
In plants animal type centrosomes are absent and they develop or nucleate from or near nuclear membranes in the opposite poles’ chromatids. Microtubule nucleation initiates at Xkip (TPX2) and such TPX2 structures have been observed in Arabidopsis and tobacco cell nuclear perinuclear region early prophase and the spindle later. In Arabidopsis TPX2 is exported from the nucleus in prophase.
The histone protein CenH3 is both necessary and sufficient to trigger the formation of centromeres and pass them on from one generation to the next; centromere contain CenH3; H3K4me2, http://www.nature.com
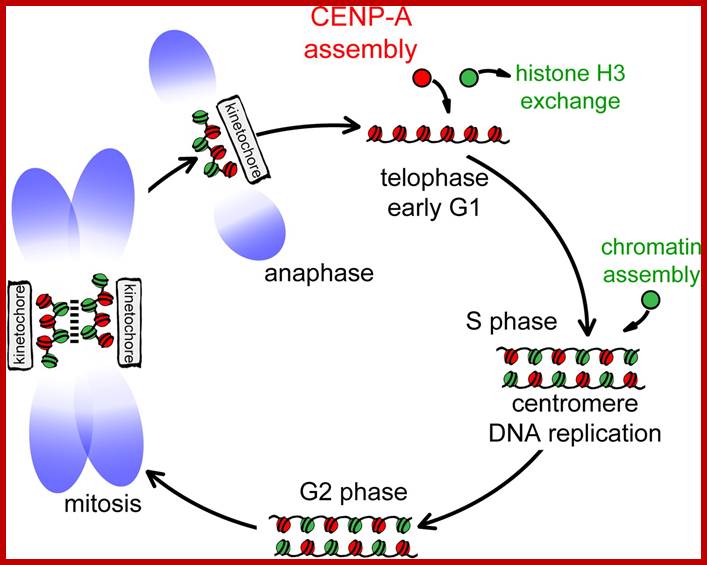
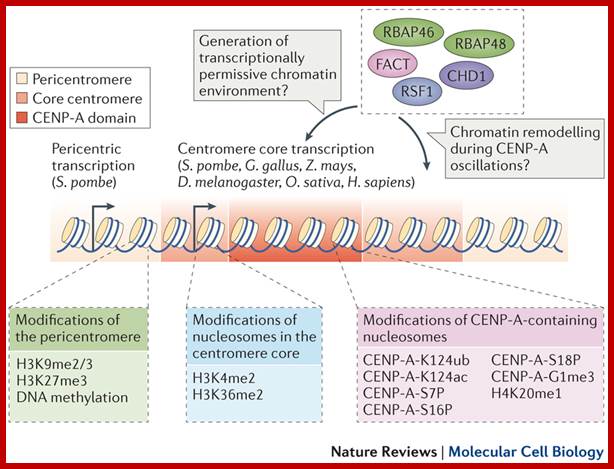
The diagram shows centromeric chromatin composition in relation to cell cycle.; http://jcb,rupress.org
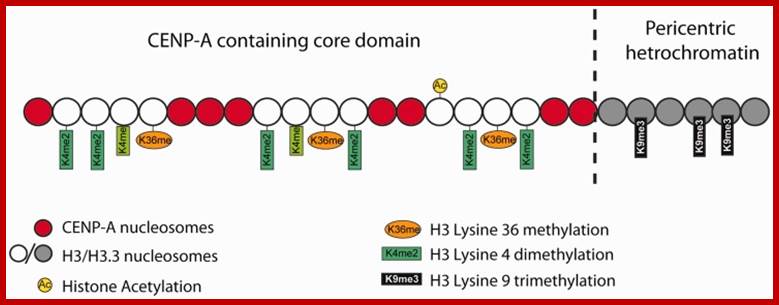
Pericentric heterochromatin consists of H3K9 trimethylation, which is vital for HP1 localization to the pericentric domains. The centromere core domain consists of clusters of CENP-A and H3 nucleosomes. In S-phase, both the canonical replication-dependent H3.1 and the replication-independent H3.3 are loaded onto centromere chromatin. The H3.1/H3.3 nucleosomes are enriched for H3K4 dimethylation and H3K36 methylation. No H3K4 trimethylation or H3K9 trimethylation could be detected at the centromere core domain. Although stretched chromatin fibre experiments indicate that histone acetylation is absent at the centromere core domain, but chromatin immunoprecipitation studies have detected a low level of H3 acetylation at the centromere core. However this histone acetylation may be tightly regulated by cell-cycle dynamics. It is currently unknown whether the histone modifications detected so far are carried by H3.1 or H.3 nucleosomes, or if the histone modifications show preferential enrichment at either H3.1 or H3.3: www.ncbi.nlm.nih
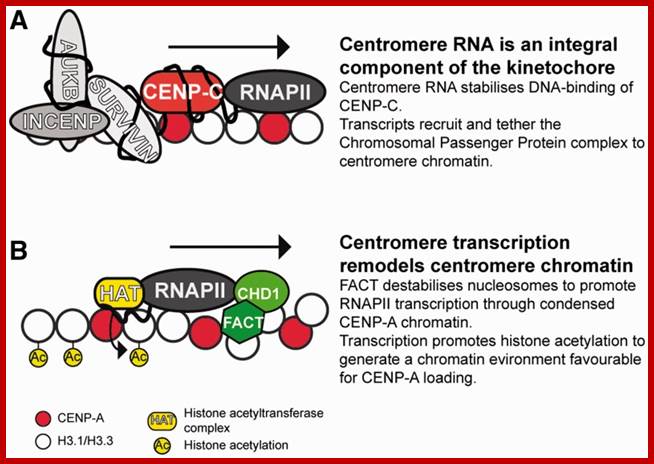
(A) Centromere RNA has been shown to associate with CENP-C and stabilize its DNA-binding ability. The localization of CENP-C has been shown to be dependent on the presence of ssRNA at the mitotic kinetochore. Centromere RNA has also been shown to associate with CPC proteins, Survivin, INCENP and to mediate the kinase activity of another CPC proteins, AUKB (70). This suggests that centromere RNA could act as a molecular scaffold at the mitotic kinetochore to recruit and organize kinetochore proteins at the centromere. (B) The act of transcription could also have an important function. The histone chaperone and chromatin remodeler, FACT complex and CHD1, has been shown to be important for CENP-A loading (90). The nucleosome destabilization activity of FACT could function to promote RNAPII transcription through the compact CENP-A chromatin, while RNAPII transcription could drive further chromatin remodeling at the centromere domain. In particular transcription could promote histone acetylation. A peak of histone acetylation has been reported to occur during mitosis (92). RNAPII transcription could recruit HAT complexes at the mitotic kinetochore to generate an acetylated chromatin environment, which has been shown to be favorable for CENP-A loading; _ Nucleic Acids Res. 2012 http://www.ncbi.nlm.nih.gov/
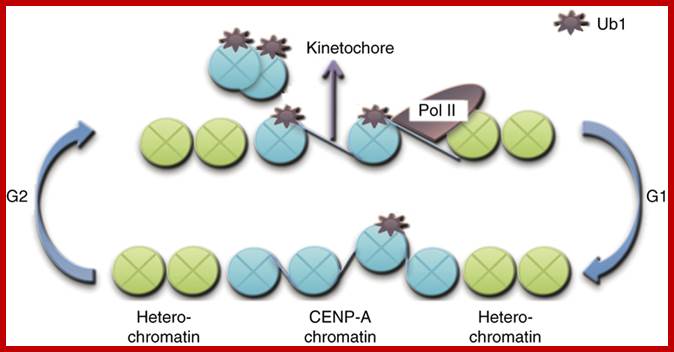
· Centromeric histone H2B monoubiquitinating promotes noncoding transcription and chromatin integrity;; Functional centromeres are essential for proper cell division. Centromeres are established largely by epigenetic processes resulting in incorporation of the histone H3 variant CENP-A. Here, we demonstrate the direct involvement of H2B monoubiquitinating, mediated by RNF20 in humans or Brl1 in Schizosaccharomyces pombe, in centromeric chromatin maintenance. Mono-ubiquinated H2B (H2Bub1) is needed for this maintenance, promoting noncoding transcription, centromere integrity and accurate chromosomal segregation. A transient pulse of centromeric H2Bub1 leads to RNA polymerase II–mediated transcription of the centromere's central domain, coupled to decreased H3 stability. H2Bub1-deficient cells have centromere cores that, despite their intact centromeric heterochromatin barriers, exhibit characteristics of heterochromatin, such as silencing histone modifications, reduced nucleosome turnover and reduced levels of transcription. In the H2Bub1-deficient cells, centromere functionality is hampered, thus resulting in unequal chromosome segregation. Therefore, centromeric H2Bub1 is essential for maintaining active centromeric chromatin. Laia Sadeghi et al
http://www.nature.com/
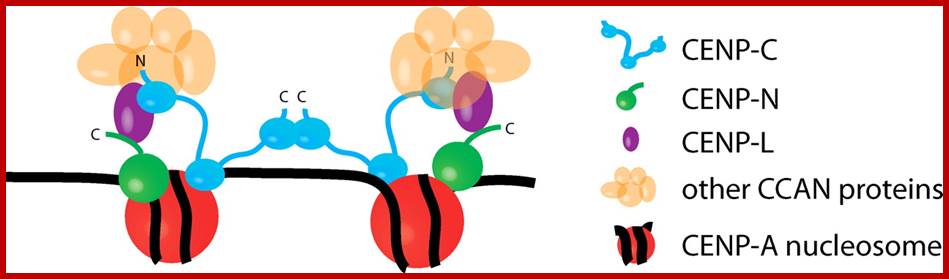
A model for centromere assembly in human cells. CENP-A (red), CENP-C (cyan), CENP-N (green), and CENP-L (magenta) nucleosomes are shown. Other CCAN proteins are colored orange. The N and C termini of CENP-C are indicated, as is the C terminus of CENP-N. http://jcb.rupress.org
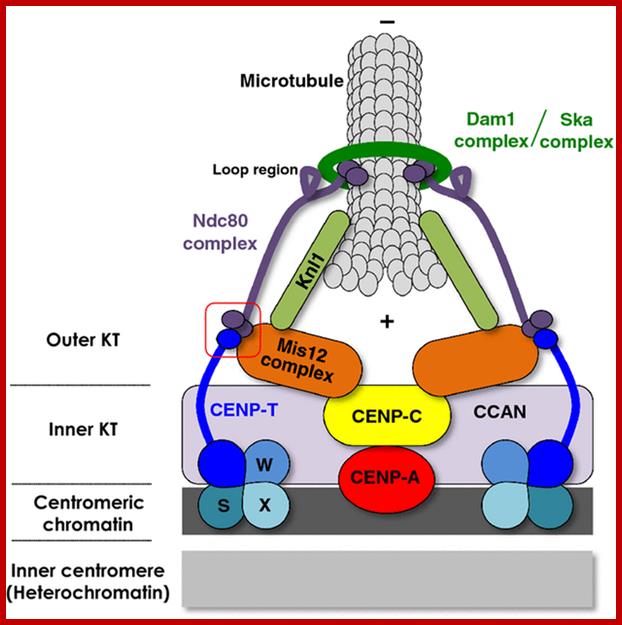
In budding yeast, the CEN DNA (centromere DNA element CDE), is recognized as CDEI, CDEII and CDEIII. Kinetochore complex is found on either side of double stranded chromatid, at CEN region and it is made up of more than 50 proteins (nearly 200) such as CBF1, CBF3 binds to CDEIII, helix loop helix binds to CDEI; they are recognized as inner kinetochore plate. The inner kinetochore interacts with Centromeric chromatin. The centromeric CEN-DNA in the chromatin, is about 0.1 to 4mb long, contain CENP-A as a nucleosome with other histones acts as a scaffold on which other kinetochore proteins assemble. The CENP-A is associated with tetrameric SX/W on either side of CENP-A. In the upper region of CENP-A another CENP-C present; on either side of CENP-C one finds CENP-T and CCAN, they form inner kinetochore complex or plate. On the dorsal surface of the CEN-C/CENP-T and CCANMis12 complex is anchored which in turn bound to plus end of Microtubule anchored by specific proteins such as Knl1- loop region of NDC80 complex encircled by Dam1 and Ska complex in the form of a ring. http://www.nature.com/

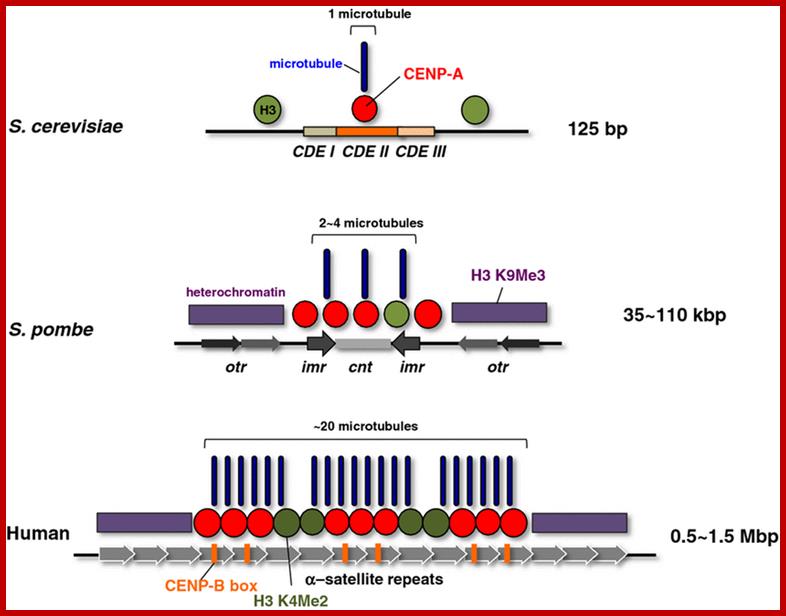
Epigenetic regulation of centromeric chromatin: old dogs, new tricks? The Centromeric region and its DNA length and the number of nucleosomes varies from one specie to the other as shown in the figure, but the constant nucleosome is CENP-A; Robin C. All shire & Gary H. Karpen; http://nature.com
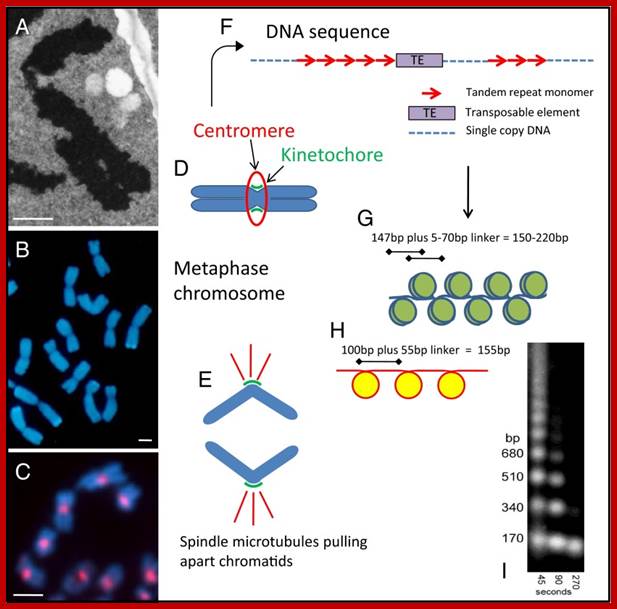
Features of centromeric DNA from different viewpoints. (A) An electron micrograph of a section of a metaphase chromosome of a wild wheat species, showing two arms with the centromere at the bend. (B) Metaphase chromosomes of triticale fluorescing blue in the light microscope. Constrictions at the centromeres are visible on each chromosome, with the two chromatids that will separate as the cell divides. (C) Chromosomes from a cell culture line of the model species Arabidopsis thaliana, labeled with a centromeric histone antibody. (D) A diagram of a metaphase chromosome showing the two arms each of two chromatids, separated at the centromere (E) and dividing into chromatids which segregate and are pulled by spindle microtubules (red) attached via the kinetochore at the centromere. (F) DNA motifs found in many centromeres, with blocks of tandemly repeated satellite DNA monomers interspersed with single copy DNA and transposable elements. (G) A diagram of the packaging of double stranded DNA (blue) into nucleosomes, with 147 bp of DNA wrapping 1.67 times around each octamer of the canonical histone proteins (olive) and fixed phase of the nucleosome within the repeat monomer. (H) The unique packaging reported by Zhang et al. (1) with ∼100 bp of the rice CentO tandem repeat sequence (red) folding once around the nucleosome core that includes CenH3 (yellow). (I) A key method for nucleosome analysis involving micrococcal nuclease digestion of chromatin and size separation of the resultant DNA fragments; the enzyme cuts DNA in the linker regions and, over the time course shown, isolates more mononucleosomes, and trims overhanging DNA not protected from digestion by the histone proteins (12). (Scale bars for A–C, 2 µm.); http://www.ncbi.nlm.nih.gov/
Centromeric structure and the size varies from one organism to the other. The length of CEN DNA can be 40kbHuman centromeres consist of α-satellite DNA arranged in tandem into higher order repeats (each arrow), and some α-satellite DNA contains CENP-B binding sites (CENP-B box). CENP-A localizes to a portion of these arrays. The number of microtubule attachment sites also varies among organisms. CENP-B is a highly conserved centromere protein in mammals and binds to a 17-bp motif in a CENP-B box. It has been shown that α-satellite DNA with a CENP-B box is responsible for de novo centromere assembly in human somatic cells (Masumoto et al.,2004)
Centromeric chromatin underlies the kinetochore which contains inner and outer plates. In mammals the centromeric chromatin is folded into nucleosomes that are modified by demethylation of lysine4 of histone H3 (H3(H3Kme2) form discrete domain of internal to CENP-A nucleosomes- found in alpha satellite DNA region (500 to 1500kb). http://www.nature.com/
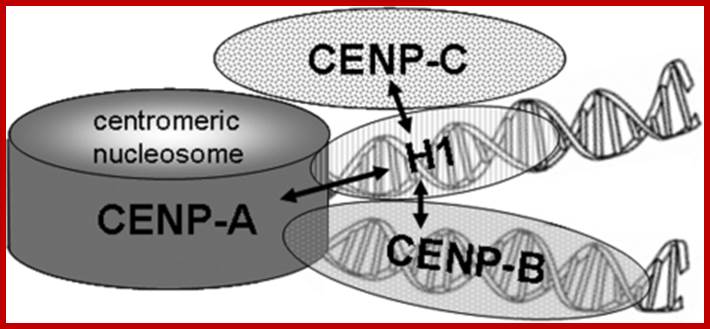
Schematic representation of centromeric chromatin with a CENP-A containing nucleosome: In this model, the internucleosomal linker DNA is alternatively occupied by histone H1 and CENP-B. The arrows indicate the protein associations detected in this study. https://openi.nlm.nih.gov/
The vertebrate kinetochore complex assembles at the centromere on alpha-satellite DNA. In humans, alpha-satellite DNA has a repeat length of 171 bp slightly longer than the DNA in the chromatosome containing the linker histone H1. The centromere-binding protein CENP-B binds specifically to alpha-satellite DNA with properties of a centromeric-linker histone. Here, we analyzed if linker histone H1 is present at or excluded from centromeric chromatin by CENP-B. By immunostaining we detected the presence, but no enrichment or depletion of five different H1 subtypes at centromeric chromatin. The binding dynamics of H1 at centromeric sites were similar to that at other locations in the genome. These dynamics did not change in CENP-B depleted cells, suggesting that CENP-B and H1 co-exist in centromeric chromatin with no or little functional overlap. By bimolecular fluorescence complementation (BiFC) and Forster resonance energy transfer (FRET), we revealed that the linker histone H1 subtypes H1 degrees and H1.2 bind to centromeric chromatin in interphase nuclei in direct neighborhood to inner kinetochore proteins.
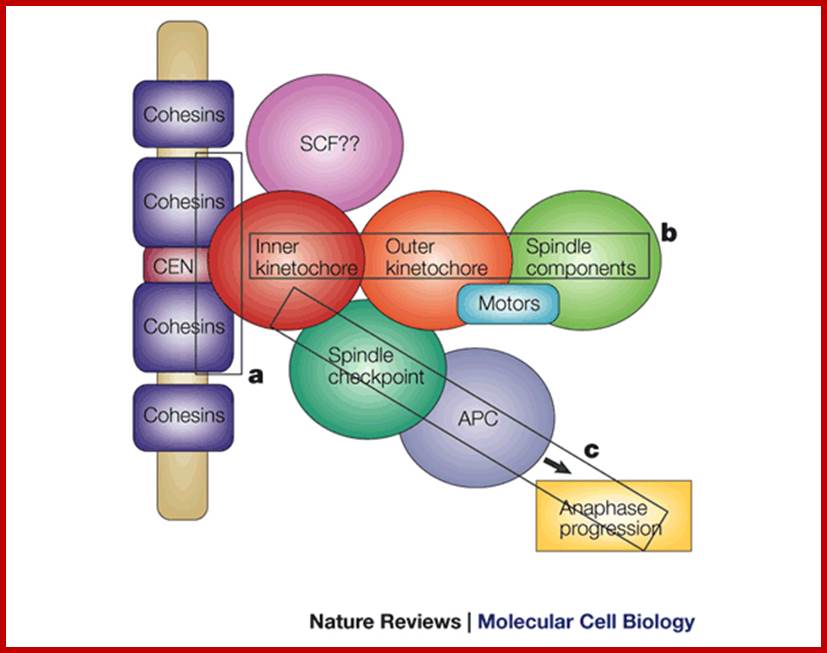
Cemntromere associated components;WWW.nature.com
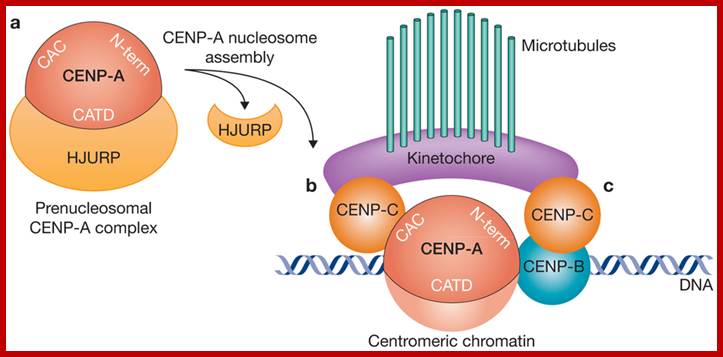
CENP-A acts as the central component for the assembly centromere and Kinetochore elements, CENP-A associates with CENP-C and CENP-C; CENP-A and CENP-C are bound to CEN DNA as nucleosomes; it is on which CENP-C bind and then Inner kinetochore associate on which Outer Kinetochore binds to which spindle fibers bind;www.nature.com
 http://femsre.oxfordjournals.org/
http://femsre.oxfordjournals.org/
Vascular plant cell division is characterized by open mitosis, during which cytoplasmic microtubules (MTs) organize into a bipolar mitotic spindle; In addition, perinuclear MTs radiating towards the cytoplasm increase in density, indicating initiation of new MT nucleation events. These microtubular arrays are completely reorganized from G2 to metaphase, starting with formation of a pro-spindle and ending with the metaphase plate, which corresponds to an equilibrium state before chromatid segregation.

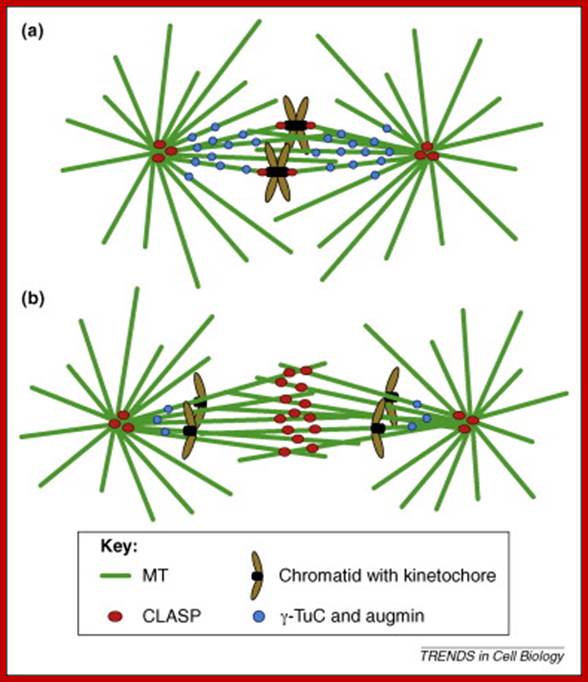
The centromere–Kinetochore region: At the heart of the kinetochore is a specialized nucleosome that contains centromere protein (CENP)-A, a histone H3 homologue48. Several inner-kinetochore components (cyan and purple ovals) associate with kinetochores throughout the cell cycle48, 174, 175. Many other proteins, including those in multiprotein complexes that contain the Ndc80/HEC1, Mtw1/MIS12, minichromosome maintenance protein-21 (Mcm21) and spindle pole component (Spc)105/KNL-1 proteins, are recruited to the outer kinetochore specifically in mitosis. They provide a landing platform for the spindle-assembly checkpoint (SAC) proteins56. The Ndc80/HEC1 complex seems to be directly involved in microtubule binding80, 81. Several microtubule-plus-end-binding proteins (+TIPs) are important for microtubule–kinetochore attachment176. Borealin (BOR), Survivin (SUR), Aurora-B (AurB), inner centromere protein (INCENP) and mitotic centromere-associated Kinesins (MCAK) preferentially populate the centromere region and regulate the stability of microtubule–kinetochore attachments. They have been implicated in the correction of attachment errors94. Several subunits of the nuclear pore complex (NPC) also localize to mitotic kinetochores, but their kinetochore function is unclear. The anaphase-promoting complex/cyclosome (APC/C) is recruited to mitotic kinetochores in a SAC-dependent manner77, 107. The ROD (rough deal)–ZW10 (zeste white-10)–ZWILCH (RZZ) complex mediates kinetochore recruitment of mitotic-arrest deficient homologue (MAD)1–MAD2 (Ref. 31). Large cytosolic pools of MAD2 and CDC20 exist besides the populations that are recruited to the kinetochore. This might be true for other SAC proteins, including budding uninhibited by benzimidazole (BUB)R1 and BUB3, and for the APC/C itself. As discussed in the main text, MAD2 adopts two conformations, O-MAD2 (open MAD2) and C-MAD2 (closed MAD2), which are depicted as red circles (O-MAD2) and yellow circles (C-MAD2). Most proteins indicated in this drawing are present at kinetochores in all metazoans. CLASP, CLIP-associating protein-1; CLIP170, cytoplasmic linker protein-170; EB1, end-binding protein-1; kMTs, kinetochore microtubules; LIS1, lissencephaly-1; MPS1, multipolar spindle-1; PLK1, polo-like kinase-1; RanBP2, Ran-binding protein-2; Ran-GAP, Ran-GTPase-activating protein; ZWINT, ZW10 interactor. https://www.researchgate.net
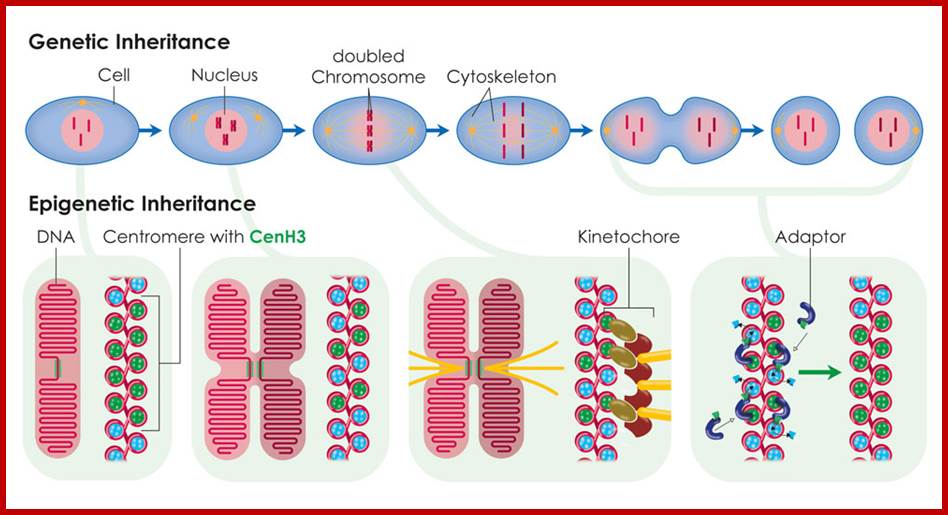
During cell division, the histone CenH3 ensures that new protein is integrated into the two DNA strands. The position of the centromere can thus be passed on from one generation to the next. ;The above diagram depicts the epigenetic inheritance of CENs; https://www.mpg.de.
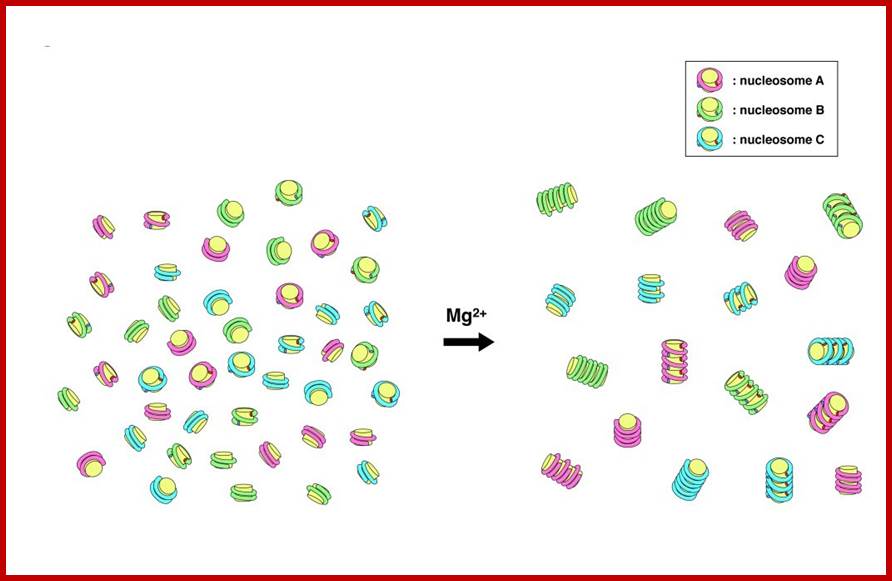
Nucleosomes have sequence-dependent self and nonself discrimination properties. This study suggests that contact points in the association of nucleosomes reside on the core histone protein and bound DNA on them; http://www.intechopen.com/
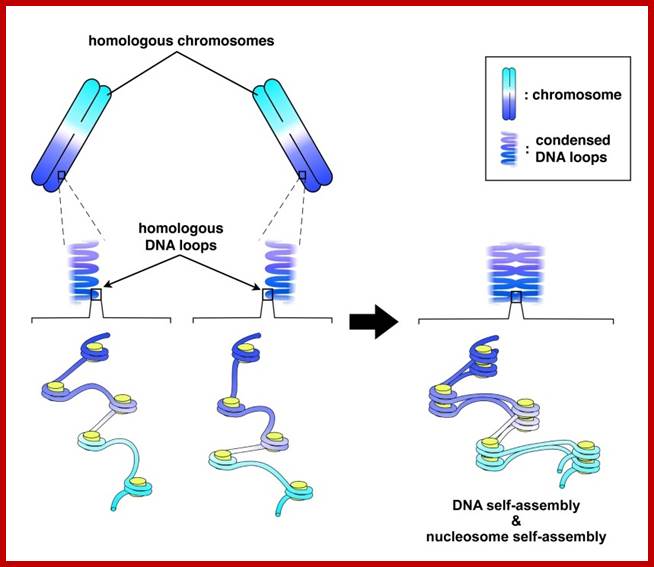
Schematic representation of DNA self-assembly and nucleosome self-assembly in paired homologous chromosomes; Both DNA and nucleosomes have sequence dependent self and non-self-discrimination properties, and can self-assemble. This property is key for presynaptic alignment that provide attractive forces facilitating DNA self -assembly and nucleosome self-assembly in meiotic chromatids (leptotene threads to pair) homologous pairing in zygotene; Schematic representation of DNA self-assembly and nucleosome self-assembly in paired homologous chromosomes. http://www.intecopen.com
Metaphase: As the nuclear membrane disintegrates into fragments in early prophase, some nuclear membrane fragment retains nuclear pore complexes; tractile fibers with continuous fibers appear in dome shaped form, which then is called mitotic apparatus. Tractile fibers appear and attach to kinetochore elements of chromatids. There is another type called polar microtubules which radiate from the polar granules. By contractile mode the tractile fibers bring all the chromosomes (which divalent i.e contain two sister chromatids, but the centromere is still intact and single. Interestingly, at CEN region kinetochore complex organizes on either side of it, thus all mitotic chromosomes contain two kinetochore complexes; they with their tractile fibers are oriented toward their respective poles. It is at the end of the metaphase the CEN region appears to be split for the kinetochores bound by their respective tractile fibers are oriented towards their respective poles. The two homologous chromosomes derived from parents when they are placed in in the equatorial region, the biparental chromosomes are oriented randomly in the equatorial region; the organization is for-2n=8
-Chromo.--Pa-Ma-Pa-Pa-Ma-Ma-Ma-Pa---
-Chromo.--Ma-Pa-Ma-Pa-Ma-Ma-Pa-Ma---
Note-Chromo- chromosomes and chromosomal nucleosomes, Ma- maternal, Pa- paternal, random orientation of parental and maternal chromatids in equatorial plane. This arrangement also holds good for meiosis parental chromosomal synapsis and segregation.
Anaphase: Once duplicated chromosomes with their chromatids placed in the mid region of the cell or what is called equatorial plate, start moving towards their respective opposite poles.
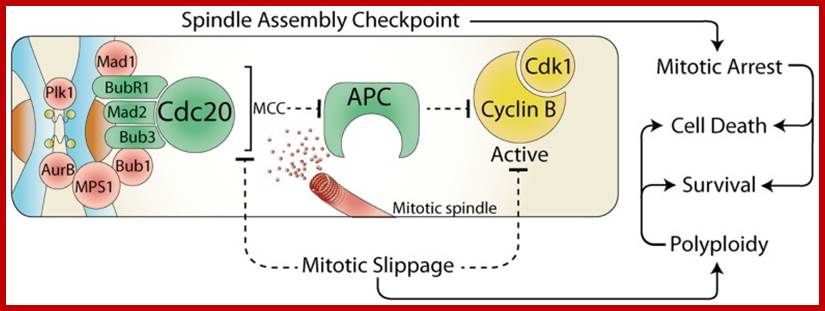
 The spindle assembly checkpoint and cell fate; during
mitosis, the constitutively active spindle assembly checkpoint (SAC) delays
anaphase until all chromosomes are attached to the mitotic spindle. Any stress
that prevents satisfaction of the SAC, results in a prolonged mitotic arrest,
which often leads to cell death. However, the SAC can be over-come by the
release of Cdc20 from the mitotic checkpoint complex (MCC) or by direct
inhibition of Cdk1. This mitotic slippage can result in polyploidy, increased
cell survival, and provides a potential mechanism for escaping mitotic cell
death. https://www.researchgate.net
The spindle assembly checkpoint and cell fate; during
mitosis, the constitutively active spindle assembly checkpoint (SAC) delays
anaphase until all chromosomes are attached to the mitotic spindle. Any stress
that prevents satisfaction of the SAC, results in a prolonged mitotic arrest,
which often leads to cell death. However, the SAC can be over-come by the
release of Cdc20 from the mitotic checkpoint complex (MCC) or by direct
inhibition of Cdk1. This mitotic slippage can result in polyploidy, increased
cell survival, and provides a potential mechanism for escaping mitotic cell
death. https://www.researchgate.net
![]()
The cell cycle is driven by the activity of specific Cyclins and specific CDKs. Cell cycle is also regulated by specific cell cycle check point proteins at G1, S and G2 and even spindle assembly check point M (SAC). The primary role SAC is to block the anaphase promoting complex (APC), which is then subjected to ubiquitination and proteasome degradation; https://www.researchgate.net
Animal cells; www.nature.com
General view not plant cells; www.cell.com
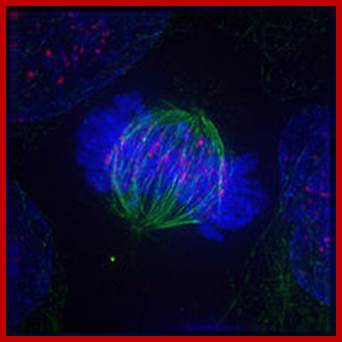
Immunofluorescent image of a cell in metaphase showing microtubules in green, chromosomes (DNA) in blue, and kinetochores in pink; https://en.wikibooks.org/wiki
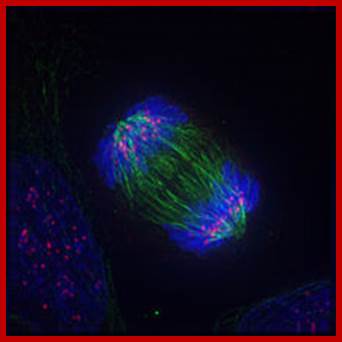
Anaphase; https://en.wikibooks.org/wiki
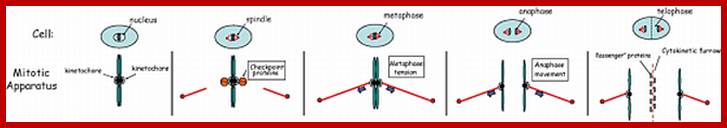
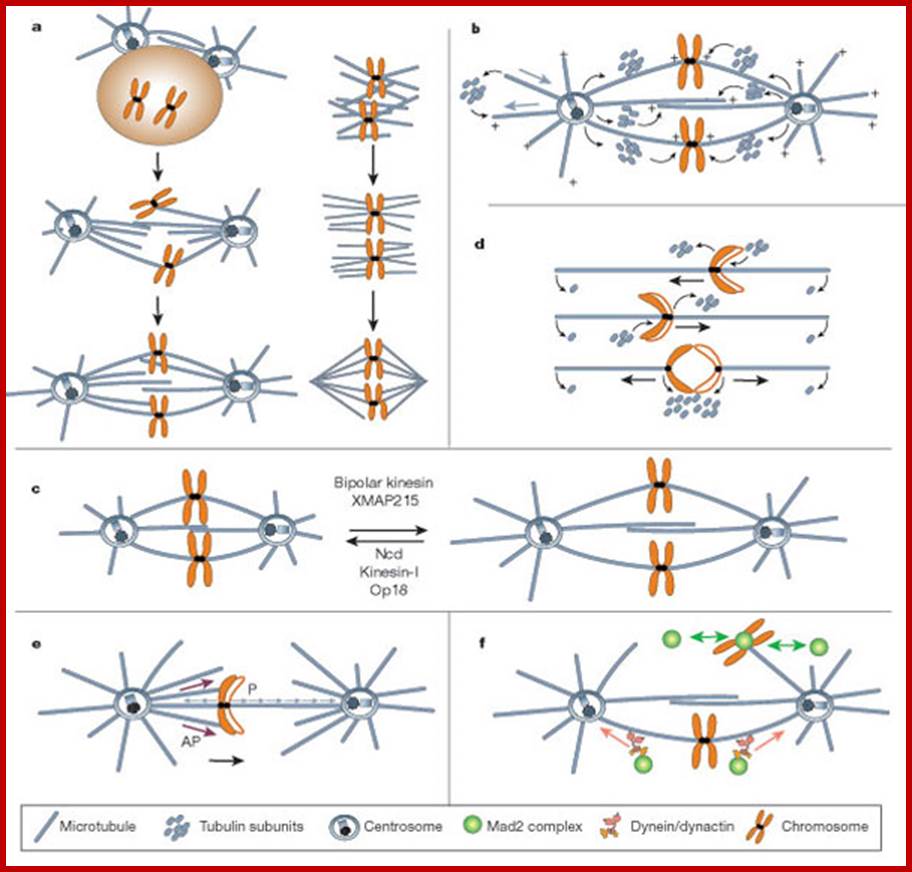
Mitotic spindle action at specific stages
In astral type cells, the beginning of the prophase is indicated by the division of centrioles and the formation of radiating fibres all-round these daughter centrioles. As the nuclear volume increases, one of the astral points starts moving towards the other pole. Thus, mitotic apparatus develops. Both in plant cells and animal cells tractile fibres which are associated with the kinetochore start depolymerizing at the poles. Thus, chromosomes move along with the tactile fibres. Perhaps, simultaneously or at a little later stage, continuous fibres grow longer with the polymerization of tubulins from the polar ends. Thus, they stretch the mitotic spindle and greatly aid in the movement of chromosomes. This process requires ATP as the energy source.
Sliding Theory: This theory envisages the presence of microfilaments associated with microtubules. The presence of Actin and Myosin units at the Polar Regions has been detected by antibodies raised against actin and myosin. Similar to that of muscular contraction, the microtubules of tactile fibres interact with acto-myosin proteins found at the poles and slide over each other. Thus, the contraction of tractile fibers towards the pole is brought about. This process also requires ATP as the energy source.
Present concept: Even though both the above said theories are attractive, each of them has their own drawbacks. It is presumed that both the mechanisms may be operating simultaneously. To begin with, the sliding mechanism starts pulling the tactile fibres at the poles, at the same time the tractile fibers undergo depolymerization at their (-) ends and continuous fibers get elongated by polymerization of added tubulins at Plus ends. Nevertheless, the knowledge about the molecular mechanisms of the organization of mitotic apparatus and their exact role in chromosomal movement and cytokinesis is far from clear.
Telophase: In this stage single stranded chromatids that are pulled towards their respective poles start aggregating; simultaneously chromosomes start decondensation. Thus, the chromosomal strands become longer. At this stage transcription activity of chromosomal DNA begins. At the same time the nuclear membrane vesicles start appearing all-round the chromosomes and soon these membranous bits with pore complexes organize into nuclear envelope. The relaxed chromatin attaches to matrix proteins at regions called MARS. As the chromosomal strands recoil and relax, the nucleolar DNA present in the region of secondary constriction loops out and nucleolar region begins to get organized. The nacked rRNA genes in clusters start transcribing precursor rRNAs. It is at the same time very many pre-rRNA processing snoRNAs and their associated proteins assemble in the nucleolus. A little later the incoming ribosomal structural proteins assemble on r.RNA and nucleolus gets organized. It is important to note that chromosomes at this stage remain single stranded.
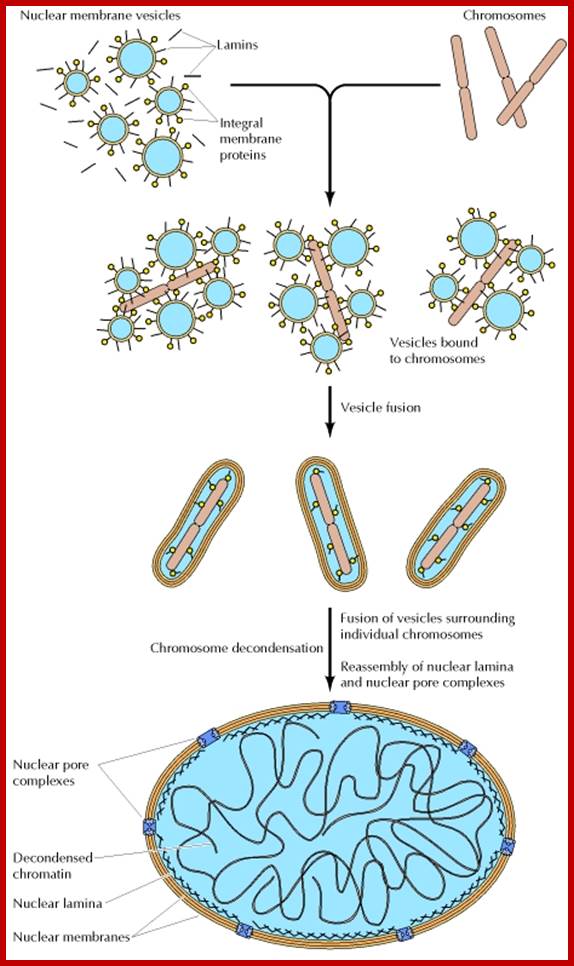
Re-formation of the nuclear envelope; The first step in reassembly of the nuclear envelope is the binding of membrane vesicles to chromosomes, which may be mediated by both integral membrane proteins and B-type lamins. The vesicles then fuse, the nuclear lamina reassembles and chromosomes get decondensed and the nucleolus gets organized on Nucleolar region of specific chromosomes.; http://www.ncbi.nlm.nih.gov/
Cytokinesis
Generally, karyokinesis leads to cytokinesis, but in certain organisms like plasmodia, siphonales algae and others, cytokinesis does not follow karyokinesis and repeated nuclear divisions lead to multinucleate cells or coenocytic cells. Similarly, during early development of liquid endosperm in coconut fruits, nuclei divide repeatedly before cytokinesis sets in. The mechanism of cytokinesis in animal cells and plant cells vary; in the former case it is achieved by cleavage and in the latter, cytokinesis takes place by phragmoplast formation.
Cytokinesis by cleavage: In animal cells, cytokinesis sets in at late anaphase or early telophase. The appearance of dense materials at the equatorial region of mitotic apparatus is the first indication of cytokinesis. In the central region tractile and other protein elements bound to membranes in the equatorial region, actin protein elements start a ring of constriction; at this point gradually a number of membranous vesicles appear at this region and then a ring of depression further deepens into a constriction or deep furrow, finally it leads to the division of cytoplasm into two units.
However, during cleavage form of cytokinesis, at the equatorial region a ring of Acto-myosin filaments appears in the cortical region of the cell. Using ATP as the source of energy these acto-myosin filaments interact. As a result, the associated protein filaments contract and the membrane to which these protein filaments are bound is drawn inwards all-round till the membranes fuse in the middle. Thus, the cells get separated. The exact mechanism of membrane contraction involving microtubules, actin and myosin and ATP is not clear. Nevertheless, the presence of these structures in and around the mitotic apparatus is known. Their involvement in the cleavage is just a presumption, of course with valid reasons.
With the development of deep constriction, some of the spindle fibres disappear due to disassembly of microtubules into tubulin monomers. Even after cleavage, some remnants of microtubules that are found at the centriole region, also disappears at the end.
Plant Cytokinesis by Phragmoplast: Plant cells which do not possess centrioles, during cytokinesis cells produce numerous membranous vesicles derived from Golgi complex and endoplasmic reticulum (SER). These vesicles appear at the inter-zonal region of the equatorial plate. Many microtubules are found at this region. With time lapse, some more microtubules are added to the peripheral mitotic spindle. Thus, the mitotic apparatus appears to be bulged. Such a bulged structure of mitotic apparatus is called phragmoplast. Later, the vesicles found in the equatorial region within the mitotic apparatus fuse with one another and form a circular membranous cisterna, which gradually extend laterally and reach the phragmoplast surface.
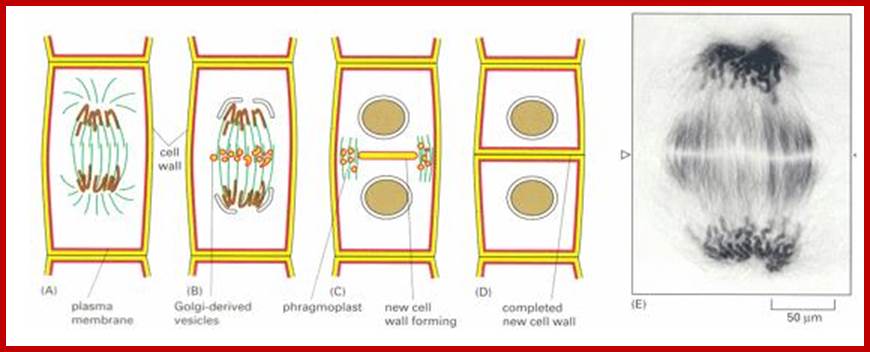
Final steps in plant cytokinesis; http://php.med.unsw.edu.au/
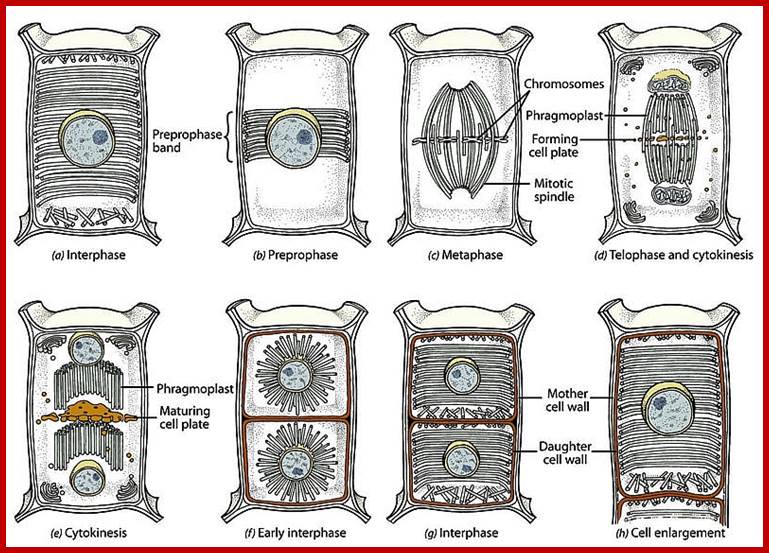
Plant cell Mitotic stages; www.searchpp.com

Cytokinesis plants; www.cell.com
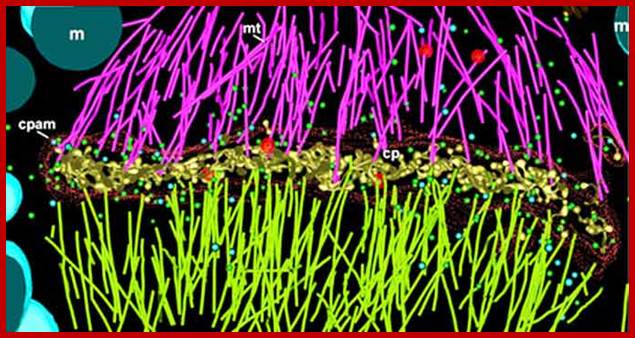
Electron tomographic view of cell plate formation:
A cell plate in the tubular-vesicular network phase of cell plate formation; Secretory vesicles (small blue and green spheres) are trafficked down the phragmoplast microtubules (light green and magenta rods, mt) to fuse with the growing cell plate (yellow, cp). A few clathrin-coated vesicles (large red spheres) can also be seen budding from more mature sections of the plate and traveling along the phragmoplast MTs. The cell plate is enclosed within a ribosome-excluding cell plate-associated matrix (red dots). The large blue structures are mitochondria (m). For clarity, the endoplasmic reticulum is not shown. Image courtesy of Dr. José Seguí-Simmaro. http://www.illuminatedcell.com/;publishing.cdlib.org
Prior to division, plant cell produces numerous vesicles containing, carries raw materials needed to create cell walls or strengthen it at the inner surface. These materials are held in an inert state, with inactive enzymes needed to form wall materials. As telophase begins, these vesicles aided by cell cytoskeletons, begin to line up down the equator of the cell, and begin to fuse (what triggers this action? will it trigger the enzymes?). As the vesicles fuse, inactive components released and they become active; cell walls begin to form, more vesicles fuse (remember the membrane is dynamic), the wall continues to grow as cell plate. Eventually the membrane surrounding middle plate will fuse with the parental membrane. Once the cell plate fuses with the parental membrane, two new daughter cells are formed. The cell plate also develops plasmodesmata for symplastic flow of liquids across. Robert Maxwell.
Finally, both the cisternae and phragmoplast reach the lateral plasma membrane and fuse with it. Thus, the cytoplasm gets divided into two compartments by the membranous cisternae which act as the newly formed plasma membranes of the daughter cells. The space found between these membranes will be soon filled up by calcium pectate which acts as the middle lamella. Then the Golgi complex derived vesicles filled with cellulose fibers and other cell wall components are directed with the help of microtubules towards the newly formed middle lamellae and cells wall materials, made up of cellulose fibres, is laid on either side of the middle cell pate. Thus, two daughter cells are produced.
Significance of Mitosis
All multicellular organisms, as well as unicellular organisms use mitosis as a mechanism for multiplication of cells. During this process, chromosomes of parental cells duplicate and distribute equally to their daughter cells. Here the term ‘equally’ denotes both quantitative as well as qualitative. This process also helps in the growth of an organism. In many organims where certain organs or cells are subjected to wear and fare, the cells are replaced continuously by mitosis, for example: in human beings there are about 2.5x1015; red blood cells and they have an average life of 420 days. In order to maintain the constancy of the blood cells, the body produces about 2.5 million new cells every second to compensate the loss, which appears to be incredible, but human body does it with mitosis.
Mitosis and Cancer (cancer is explained as separate topic)
Even though mitosis helps in the growth of an organism not only in the size but also in population, it is a highly related phenomenon. By mutational studies in yeast cells, as many as 38 or more steps have been identified to take part in mitosis, of which some are highly crucial in the progression of mitotic stages. If there are any mutations in the genome that control this process, cell division is completely inhibited or completely goes out of order or it may end up in an uncontrolled mitotic division. Under normal conditions, particularly in multicellular organisms, mitotically derived cells undergo differentiation and perform specific functions. Instead, in an uncontrolled process, cells undergo continuous multiplication by repeated mitosis. In these cases, the cell derivatives do not undergo any differentiation, but they divide and redivide endlessly. As a consequence of this, innumerable cells of the same kind are formed. Such a group of cells which are endowed with a potentiality to divide and redivide ceaselessly is called tumor cells and the disease thus produced is referred to as cancer. This can be induced by various carcinogenic agents like drugs, X-ray irradiations and even some viruses. Certain spontaneous mutations may also cause growth.
The analysis of cancer cells indicate that the rapid and uncontrolled cellular divisions are due to some changes in the regulatory chromosomal proteins called non-histones. Identification of such causative non-histones is very essential and important to cure the cancer disease.
In plants, however, callusing or callus formation is another example of uncontrolled, undifferentiated tumor formation. Nevertheless, the callus formation is known to be controlled by certain phytohormones like auxins. The special feature of these hormones is that at particular concentration, they induce tumor formation in plant cells, but at a different concentration with other hormones like cytokinins, they may induce differentiation of shoots or roots. The probable mechanism by which the hormones cause callus formation is again attributed to differential gene expressions or due to certain modifications of nonhistone proteins, which actually trigger off the cellular components to undergo such uncontrolled cell divisions.
Mitosis and cloning
Development of a multicellular organism always begins with the zygote which is nothing but the product of syngamy. The zygote inturn undergoes repeated but controlled cell divisions which are followed by cell differentiation, where the cell derivatives develop into different types of cells which have their own characteristic structures and functions. The overall growth of an organism thus depends upon a controlled, determinate cell division and differentiation. The molecular basis of such cell differentiation is not clear, though certain differential gene expressions in E. coli, Drosophila and others have been very well studied.
Using the property of cell’s totipotency, where a single cell could be induced to develop into a complete organism, biologists have succeeded in the clonal propagation of plant in general and animals in specific cases. Normally, the production of off springs involves sexual reproduction, where two parents contribute the gene pool through gametes. Such offsprings possess the mixture of genes from their parents. Instead, if diploid cells of one of the parents are induced to develop into an offspring, then such offsprings are referred to as clones. Such clonal propagation is in vogue, particularly in plants, where the technique of tissue culture has been very well exploited.
In this process, a cell or a group of cells from any part of the plant body is explanted into a known solid or liquid agar based nutrient medium. If the medium is appropriate and balanced with the required phytohormones, the cell or cells explanted develop into callus, from which numerous embryos can be induced at will. Later, the embryos can be cultivated. This method has been successfully employed in cultivating horticultural plants, crop plants and also plants which are difficult to multiply by vegetative propagation or by sexual reproduction.
It is important to note, that this process has employed mitosis as the most important mechanism for cell multiplication. Nevertheless, this process of cell division is always followed or preceded by a regulated differentiation. Inspite of recent technical innovations, the molecular mechanism of differentiation is not known.
Unlike plants, clonal propagation of animals has been successful only in certain cases like frogs and rats. In these cases, diploid cells from the somatic tissues, rather than germinal cells, have been successfully used. Either by the transplantation of the somatic cell into the mother uterus, or by the transplantation of a nucleus taken out from the somatic cell into enucleated zygotic cell, complete animals have been grown in the laboratories. Such animals have the genome of only one parent and such offspring’s are called clones. Though clonal propagation of human beings has been attempted, the moral, social and ethical problems have deterred him from doing any further experiments. Nevertheless, with the time and change of attitude towards the fellow human beings, perhaps, one day he may resort to such clonal propagation of man to preserve himself.
Even though clonal propagation of higher plants and animals has its own implications as well as limitations, cloning of genes by Genetic Engineering techniques has been the craze of the day; its application in the welfare of fellow human beings is unlimited. Many genetic engineering industries have been set up in USA and other European countries. The trials and tribulations in developing this elegant but sophisticated technique are unsurpassed in the recent history of molecular biology. The pace of development in this field is phenomenal and it can be compared only to the space and computer technology. Biologist have already succeeded in cloning of genes for insulin, growth hormone, interferon and work is in progress to clone nitrogen fixing genes (nif genes) into eukaryotic plants. Hitherto, man has relayed on specific organisms as the source of gene products, unfortunately the labour, time and money spent to extract them was exorbitant. Added to this, the recovery was extremely poor. But the cloning techniques have made life easy and these products can be synthesized on a large scale, thus the cost of production as come down which is a great boon for common man.
MEIOSIS:
Plants like ferns and sweet pea reproduce by spores and gametes respectively. Higher (higher) animals produce gametes as one of the modes of reproduction. Most of the above said cases and other innumerable organisms, the plant or the animal body is diploid (2n), such organisms resort to sexual reproduction by means of haploid gametes with gene recombination during meiosis. Among the lower animal and plant kingdom one finds them as haploids. Haploids use asexual mode of reproduction, but also employ sexual reproduction by the gamete fusion and produce haploid spores.
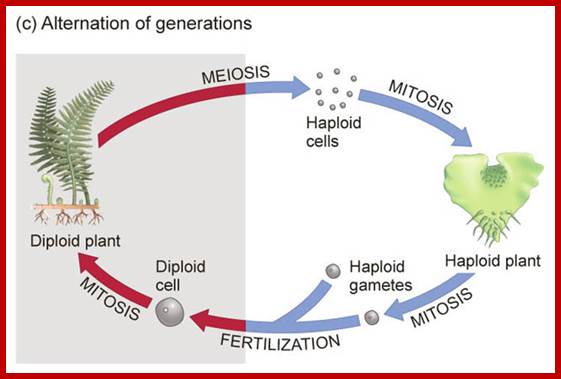
Ferns; Meiosis in Ferns during spore formation;
https://www.studyblue.com
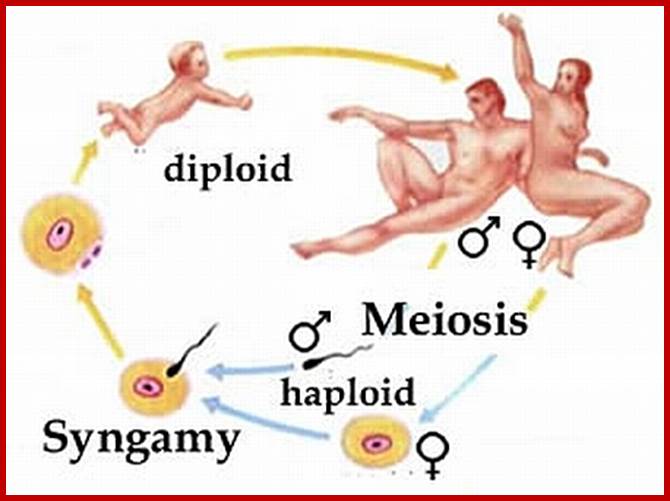
Meiotic cycle at specific stages; www.mysearch.org.uk
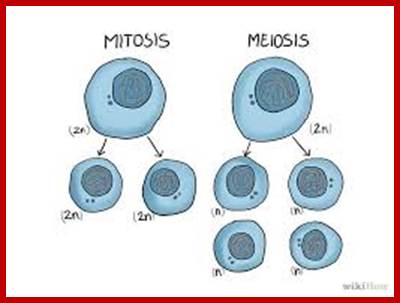
www.wikihow.com www.pinterest.com
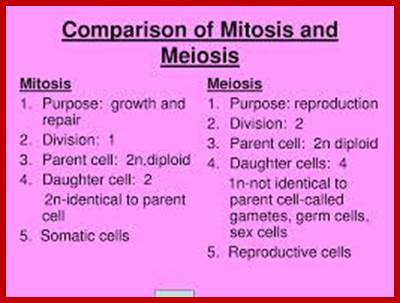
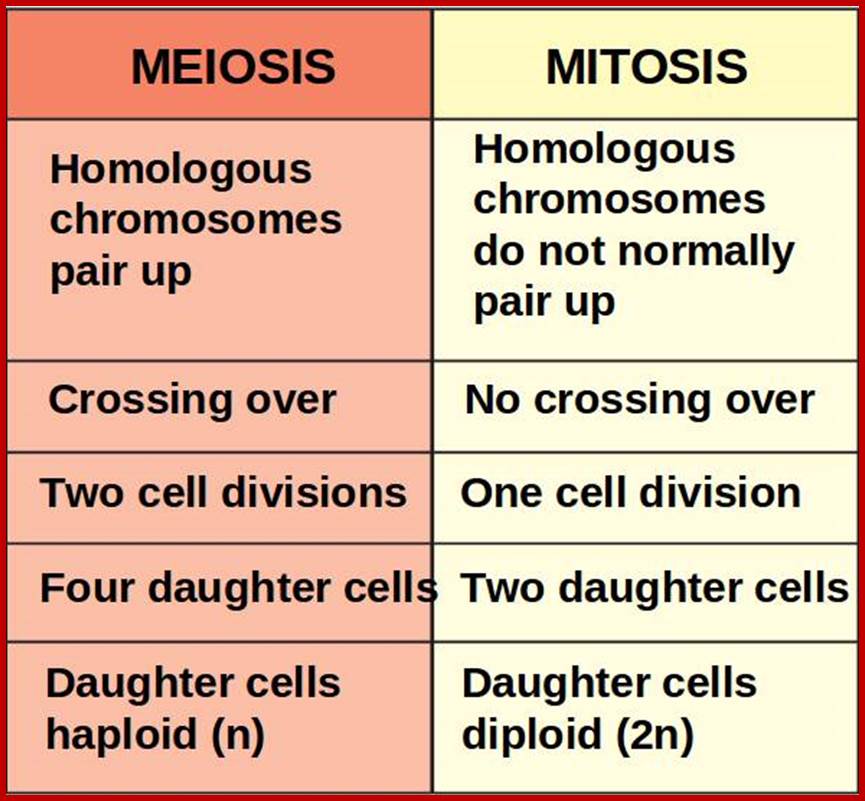
Not all cells in the body of an organism produce haploid gametes or spores. These gametes are produced only in specialized structures called reproductive organs such as gonads and testicles in higher animals, ovary-ovules and stamens-anthers in angiosperm plants both are specialized organs; there germ line cells mature with time and some are transformed into gamete producing cells and undergo meiosis and generate haploid cells, which are transformed into sperms in males and eggs in females. The transformation of diploid mother cells undergoes a specialized cell division where the diploid chromosomes are reduced to haploid set; this remarkable and unique cell division is called Meiosis. In this process diploid chromosomes with their homologous pairs, undergo synapsis and recombine and segregate and separate into haploid sets (parental). This is a unique mechanism developed over million years ago when organisms both unicellular and multicellular developed mode of asexual reproduction then they developed sexual reproduction. Haploid organisms also produce gametes and they fusion to generate their next haploid generation. The diploids require haploid gametes in both plants and animals. Sexual reproduction provides recombination and variations among the progeny.
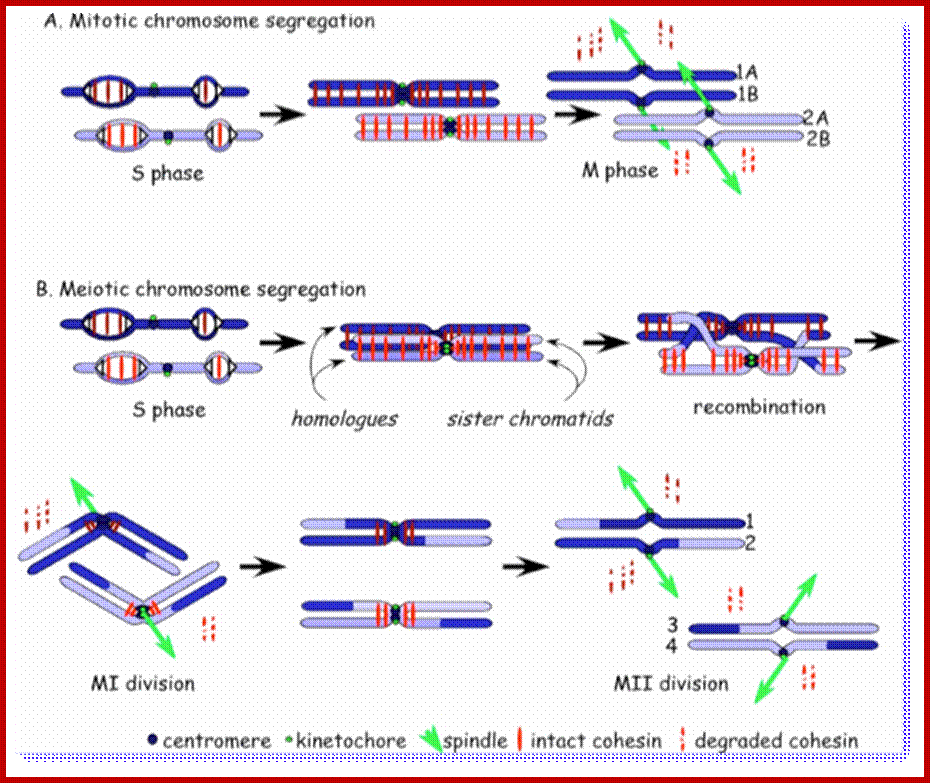
Comparison of chromosome dynamics in meiosis and mitosis: A) Mitosis in a cell with two chromosomes ensures that each daughter cell receives a copy of each chromosome. Importantly, while the apparatus of mitosis ensures that each daughter cell will have a copy of chromosome 1 and chromosome 2, it does not distinguish which one. That is, the daughters may end up with (1A and 2A), and (1B and 2B). Or, they may end up with (1A and 2B) and (1B and 2A). Since the sister chromatids are identical, this random orientation doesn't matter. The important thing is that the daughter cells have the exact same chromosome complement as the starting cell: one copy of chromosome 1 and one copy of chromosome 2. (B) In meiosis, the starting diploid is reduced to four haploids. The homologous chromosomes are duplicated, and paired to one another. After recombination, the homologues separate in the MI division. The MII division separates the sister chromatids, similar to mitosis. Each daughter nucleus will receive a single chromatid from a single homologue; importantly, because of recombination, the four daughter nuclei will not be genetically identical. Meiosis Metaphase1 and Anaphase1; segregation-http://www-bcf.usc.edu/
The gametes like sperms and eggs fuse to produce a zygote which is the first cell generation of a new offspring. For example: human body is made up of diploid cells approximately 10^14 cells. The reproductive organs are also diploid. If the cells of these germ lines produce gametes of diploid nature, the fertilized product will be tetraploid. And in successive generations the ploidy level goes on increasing. But this does not happen; the offsprings of the diploid parents will be always diploid. This is achieved by a remarkable process called Meiosis a specialized mode of cell division. The diploid reproductive cells produce haploid gametes by Meiosis, and such haploid gametes fuse to produce diploid off springs thus the diploid chromosomal number is maintained between parents and off springs by successive meiosis and fertilization. Thus, Meiosis is often considered as an antithesis for fertilization.
Genetics of Meiosis:
Meiosis unlike mitosis takes place in sporulation (in some) or gamete producing cells. Most of the cells which are set to undergo meiotic division are quite large, distinct, and rich in cytoplasm and possess large nucleus with a conspicuous nucleolus. The cells show high rate of metabolic activity. The duration of meiosis varies from organism to organism and from few hours to many days; it is waiting for the act. It takes place in two successive stages (1) Meiosis I or Reduction stage (2) Meiosis II or equatorial division stage. Each of these stages further, shows sub stages like Interphase, Prophase, Metaphase, Anaphase, Telophase and Cytokinesis.
Meiosis mode of gamete production requires 90 or more genes in Arabidopsis, but the role of 50 of them has been studied. By profiling gene expression in the mouse fetal ovary in mutants with whole tissue and single-cell techniques, we identified 104 genes expressed specifically in pre-meiotic to pachytene germcells Meiotic transcriptomes in plants have been studied in Arabidopsis thaliana, rice (Oryza sativa), wheat (Triticum aestivum), petunia (Petunia hybrida), sunflower (Helianthus annuus), and maize (Zea mays). Studies in all organisms, but particularly in plants, indicate that a very large number of genes are expressed during meiosis, though relatively few of them seem to be required for the completion of meiosis.
In Arabidopsis meiocytes, approximately 20,000 genes are transcriptionally active ; These genes constitute roughly 60% of the annotated genes in this species. In maize, about 50% of the 32,500 annotated genes are transcriptionally active during meiosis. Roughly 1,000 transposable elements, 32.5% of all transposable elements annotated genome-wide, are expressed in Arabidopsis meiocytes; Transposable elements belonging to the Copia, Gypsy, and SINE families exhibit the highest meiotic activity.
In rice, microarray studies identified 2155 genes expressed at higher levels in meiotic anthers compared to seedlings. Many of these genes have not been previously linked to meiosis A microarray study in petunia also found several novel meiotic genes; roughly 1,000 transposable elements, 32.5% of all transposable elements annotated genome-wide, are found to be expressed in Arabidopsis meiocytes; http://journals.plos.org/plosgenetics/;
Biogenesis of these secondary siRNAs is triggered by microRNAs (miRNAs). The siRNAs are either 21 ntds or 24 ntds in size and produced in a phased manner, and hence named phased secondary siRNAs phasi-RNAs; ( Song et al., 2011). In addition to these small RNAs, several miRNAs known to be involved in transcriptional gene silencing have been detected in the meiosis RNA-seq studies of Arabidopsis (Mi et al., 2008; Yang et al., 2011).
Involvement of small RNAs in meiosis has also been documented outside of plants. PIWI proteins are small-RNA-binding proteins in the germline of metazoans that belong to the family of Argonaute (AGO) proteins. PiRNAs, small RNAs associated with PIWI, serve to silence transposons in the mouse male germline. Little is known about what specifically phasiRNAs and miRNAs might do in meiosis in plants, and functional studies are needed in this area.
Studies on C.elegans, Drosophila melanogaster, Mus musculus, Arabidopsis thaliana and Oryza sativa (the last two are plants), in their specific reproductive niche germ line cells derived from stem cells by mitotic divisions enter into meiotic cell cycle and produce sperms and eggs. Poor nutrition conditions trigger the 2n cell to enter into meiosis. Such stem cells can be induced to go into meiosis by depriving nutrients. As germ cells mature, the mitotic germ cells enter into meiotic phase, which begins with “meiotic S-phase” where the genome gets replicated and meiosis specific cohesin complexes are loaded onto chromosomes.
http://www.ncbi.nlm.nih.gov/
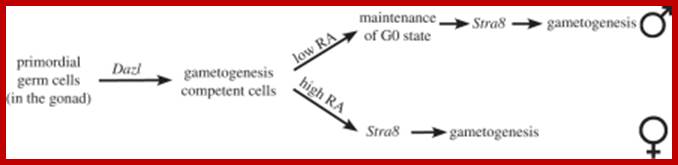
Schematic picture in the diagram above shows how germ cell fate is established in mice; cell intrinsic (DAZL activity) and extrinsic (RA) signals converge to regulate Stra8expression. The putative transcription factor Stra8 could be viewed as the functional counterpart ofIME1 and ste11+ in mammals. It is essential for gamete formation in males and females. Mice-lacking Stra8 do not enter meiosis and gametogenesis. Importantly, Stra8 expression appears to function as an integrator of internal and external cues to induce gametogenesis.
Some striking similarities between yeast and animal systems exist. In all three species, a master regulator IME 1-6 are essential for the entry into gametogenesis: IME1 in budding yeast, ste11+ in fission yeast and Stra8 in mice. Cell intrinsic and extrinsic signals converge at their promoters to regulate their expression. Cell intrinsic signals such as mating-type information and respiration in the yeasts and germ cell-specific expression of Dazl in the mouse create a gametogenesis competency state on which extracellular signals act to induce gamete formation. In S. pombe and S. cerevisiae, these extracellular signals are nutritional signals that function through highly conserved pathways—the PKA and the TOR pathways—to induce sporulation. It will be interesting to determine whether the two pathways also regulate entry into gametogenesis in mammals. Down regulation of PKA activity is required during oocyte maturation, and perhaps it is also needed during entry into gametogenesis. A recent study showed that down regulation of mTORC1 is important for maintenance of the spermatogonial progenitor cell’s population. Perhaps reduced mTOR activity is also needed for initiation of gametogenesis. Many internal and external factors and the mechanism of diploid cells enter into meiotic state is yet to be resolved.
The C. elegans mitosis/meiosis decision: (A) The nematode germline possesses a GSC pool within its somatic niche. Germ cells in the mitotic cell cycle (yellow) extend beyond the niche and include transit-amplifying (TA) germ cells, which have been triggered to begin maturation toward meiotic entry (green arrow); germ cells in the meiotic cell cycle (green) are more proximal. Overt entry into the meiotic cell cycle (red arrow) occurs asynchronously. Conventions are as in Figure 1. (B) Molecular regulation of the C. elegans mitosis/meiosis decision includes Notch signaling from the niche and FBF maintenance of GSCs, including repression of the meiotic program. Arrows indicate positive regulation; barred lines indicate negative regulation. Solid arrows and lines indicate direct molecular regulation: Notch signaling directly activates fbf-2 transcription and FBF directly represses mRNAs of the meiotic program as well as key differentiation regulators; http://www.ncbi.nlm.nih.gov/
The Drosophila mitosis/meiosis decision: (A) The Drosophila germ line possesses asymmetrically dividing GSCs (Germ line Stem Cells) within its somatic niche; transit-amplifying divisions form a 16-cell cyst and meiotic entry (red arrow) occurs in that 16-cell cyst. In males, all 16 germ cells enter the meiotic cell cycle, as depicted here; in females, only two of the 16 germ cells enter the meiotic cell cycle (not shown). Conventions and acronyms are as in Figures 1 and and2.2. (B) Molecular regulation of the Drosophila mitosis/meiosis decision relies on extrinsic BMP (Bone Morphogenic Protein) signaling and intrinsic Bam/bgcn RNA regulation, with abundant Bam controlling the position of meiotic entry. Solid arrows and lines indicate direct molecular regulation; dashed line indicates control that may be either direct or indirect. Germ-line stem cells (GSCs); Bam and Bgcn are both RBPs that have also been shown to repress mei-P26 expression. http://www.ncbi.nlm.nih.gov/
In budding yeast cells, multiple cascades of events regulate the transcription of IME1 (Inducer of MEiosis) a transcription factor (master regulator of meiosis) gets activated and acts in late G1 or at G1/S transition to activate the transcription of early meiotic genes to start meiotic S-phase; the induced genes are serine/threonine protein kinases. IME1, a key inducer of meiosis, is rapidly up regulated when diploid cells are transferred to sporulation medium. IME1 encodes a transcriptional activator recruited to promoters of early meiosis-specific genes by association with the DNA-binding protein, Ume6. Two of the early meiosis specific genes, a transcriptional activator, Ndt80, and a CDK2 homologue, Ime2, are required for the transcription of middle meiosis-specific genes that are involved with nuclear division and spore formation. Similarly, A-MYB (MYLB1) transcription factor is a master regulator of meiosis in male cells.
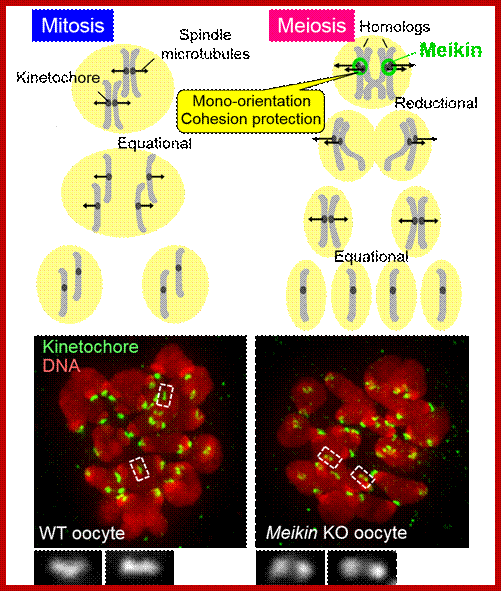
It is known that during this reductional division the kinetochores of the duplicated chromosomes are captured only from one spindle (“mono-orientation”) and remain attached throughout anaphase. Although the existence of a master regulator of meiotic chromosome segregation has been anticipated, its identity has remained elusive. The research group of Project Researcher Jihye Kim and Professor Yoshinori Watanabe at the University of Tokyo Institute of Molecular and Cellular Biosciences identified a novel kinetochore protein in mouse germ cells, the function of which was completely unknown, and which the group has named Meikin (meiosis-specific kinetochore protein). Thus, the researchers conclude that Meikin is the long-awaited master regulator of meiotic chromosome segregation. In addition, the researchers showed from an analysis of mice and yeast that the mechanism of chromosome separation in germ cells is conserved in many living organisms including humans; Meikin © 2015 Yoshinori Watanabe; http://www.u-tokyo.ac.jp/en/utokyo-research
In plants, there are specific internal regulatory differences between mitosis and meiosis however the entry and progression through meiosis are controlled by many of the same regulators as in mitosis, among many others i.e., CDK-cyclin complexes and the anaphase-promoting complex/cyclosome (APC/C) are important. In flowering plants, however, meiotic cells, i.e., megaspore mother cells (by definition the female) and microspore mother cells (male), are formed late during development; inner sub-epidermal cells of anthers and ovules undergo transformation into microspore (MiMcells) and macrospore mother cells (MaMcells).
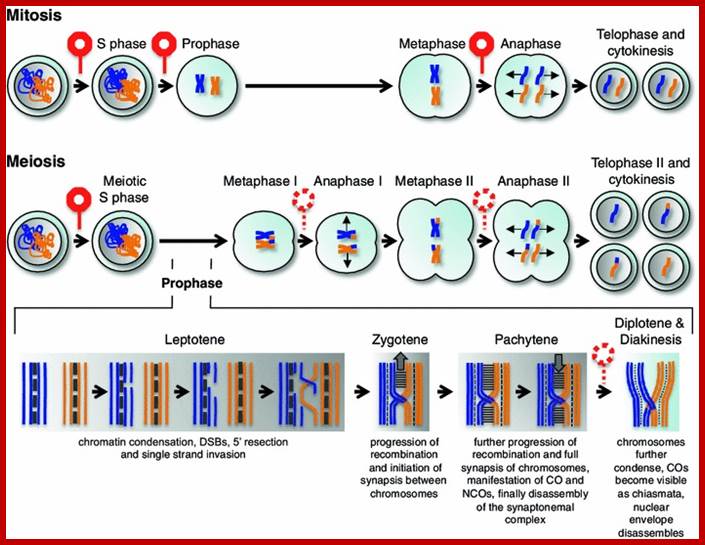
Overview of a mitotic and meiotic division. Top panel major transitions in the mitotic cell cycle. Only one pair of homologous chromosomes is shown in orange and blue, with each line representing one chromatid. Chromatids duplicate during S phase, condense at prophase and segregate at anaphase followed by decondensation. Note the absence of the nuclear envelope during mitosis. The middle panel concurrent meiotic stages, with the first meiotic division added onto the mitotic program. Note that meiosis I is unique in segregating homologous chromosomes instead of chromatids. The segregation of sister chromatids at anaphase II resembles a mitotic division. The lower panel highlights different stages of the meiotic prophase; the events at the recombination sites are largely simplified, for a more detailed description see other reviews on this topic (Edlinger and Schlogelhofer 2011; Osman et al. 2011). Please note that the leptotene stage shows the highest level of magnification, zygotene/pachytene is intermediate and diplotene/diakinesis shows the lowest magnification. Single blue and orange lines in this panel indicate single DNA strands, and two adjacent lines represent one chromatid. Double-strand breaks (DSBs) in leptotene comprise the first steps of homologous recombination. Three mitotic checkpoints are highlighted with red signs. Meiosis in plants presumably shares one checkpoint at the beginning of meiotic S phase with the one found in animals and yeast (in red), whereas other meiotic checkpoints known from animals and yeast appear to be not present or function in a relaxed manner in plants (signs in red dashed lines); www.ncbi.nlm,nih.gov
“In the case of the megaspore mother cell, this pathway includes the nuclear-localized protein SPOROCYTELESS/NOZZLE (SPL/NZZ) , WUSCHEL (WUS) a TF, activate WINDHOSE 1 (WIH1) und WIH2 ; possible receptors for the WIH peptides are the tetra-spanning-type transmembrane protein TORNADO 2 (TRN2) and the leucine-rich repeat protein TRN1. In Arabidopsis, maize and rice implicated small RNAs in regulating meiotic progression, the repression of germ cell fate in somatic tissues, or, as was shown in rice, by repressing a somatic fate in germ cells. Posttranscriptional regulation for megaspore mother cell fate specification has been further underlined by the identification of the SUPPRESSOR OF GENE SILENCING 3 (SGS3) and of RNA-DEPENDENT RNA POLYMERASE 6 (RDR6).
MEIOSIS ARRESTED AT LEPTOTENE 2 (MEL2) in rice: DNA methyl transferases in maize leads to unreduced gametes and multiple embryo sacs, exemplifying control at the chromatin level of both female and male meiosis. AMEIOTIC 1 (AM1); CDK-A and CYCLIN-A- the CDK-cyclin activity has to reach a certain threshold level. Then, a higher level of kinase activity is required for a cell to move from gap phase 2 (G2) or M phase that follows G1 to S phase in mitosis (M phase).
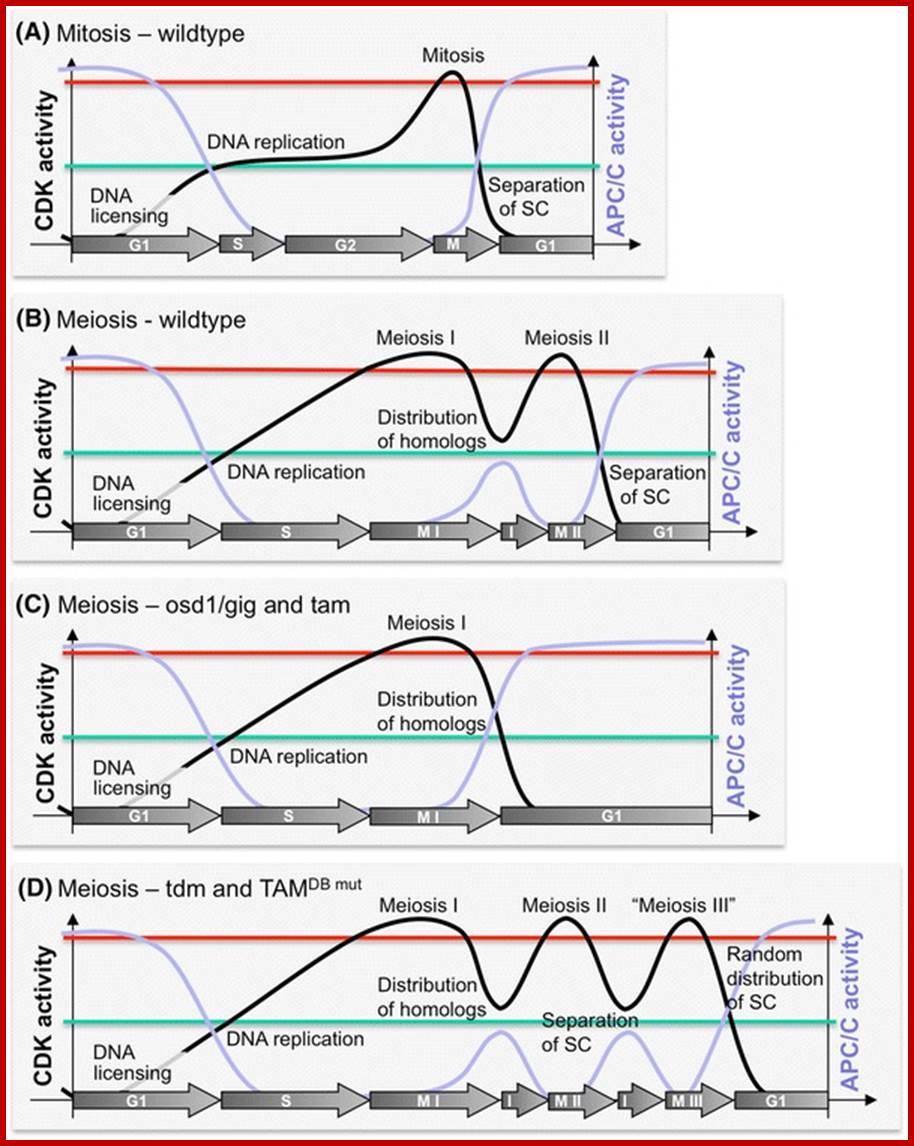
Hypothetical activity levels of CDK and APC/C complexes during mitosis and meiosis. a Progression through mitosis is thought to rely on increasing levels of CDK activity (black line). Medium levels of CDK activity are required for the induction of S phase, and high levels are necessary to promote M phase. Putative threshold levels for S phase are indicated by a horizontal green line, threshold concentrations for M phase by a red line. Please note that most likely CDK activity in plants is separated into S phase CDK-cyclin levels and M phase CDK-cyclin levels that are for simplicity reasons not separately shown here. In order to license the origins of replication for S phase, CDK activity as to be low. This is largely accomplished by the activity of the APC/C (indigo line) that mediates the degradation of cyclins at the end of mitosis and thus sets back CDK activity. APC/CCDC20 requires phosphorylation by CDK-cyclin complexes for activity but is kept largely inactive until anaphase. This inhibition will only be released if all chromosomes are attached to the mitotic spindle. The APC/C mediates then the degradation of securin which liberates separase that in turn cleaves the centromeric cohesions between sister chromatids (SC) to allow their subsequent segregation. After degradation of cyclins and drop of CDK activity, the APC/C is kept active by the Cdh1/Fzr/CCS52 adaptor protein. b During the meiotic S phase that typically takes much longer than a mitotic S phase, chromosomes are prepared for meiosis, for instance by the incorporation of the meiosis-specific cohesion REC8. Prophase I immediately starts after S phase (see also Fig. 1) that again typically takes much longer than the mitotic prophase. Dampening of APC/C activity and/or maintenance of CDK activity after anaphase I is crucial to prevent exit from meiosis and to establish interkinesis (the short phase between meiosis I and II) before meiosis II. To what level CDK and APC/C activities are changed is purely speculative in the graph. c The second meiotic division is skipped in mutants like osd1/gig and tam. Presumably, loss of TAM directly reduces CDK activity levels, while loss of OSD1 leads to full activation of the APC/C and hence a drop in CDK activity via degradation of meiotic cyclins. d Mutants in TDM and plants expressing a TAM mutant version in which the recognition sequence for the APC/C (destruction box) is mutated enter a third meiotic division in which then the sister chromatids are randomly distributed. It is plausible that such a third division, similar to the first and second division, is guided by raising and falling levels of CDK and APC/C activities. Mutants in TAM also slow down the progression of meiotic Prophase I, a feature that is not covered here; http://www.ncbi.nlm.nih.gov/
Central cell cycle in plants contain variety of CDKs (CDKA;1, CDKB1;1, CDKB1;2, CDKB2;1 and CDKB2;2), CDK-A and CDK-B1 and B2 , more than Cyclins called Cyclin-As in Arabidopsis.
A first prominent checkpoint guards the entry into S phase (G1-S transition point) and requires the activity of S-phase-specific CDK-cyclin complexes exceed a threshold level. A second checkpoint controls the entry into mitosis (G2-M transition point) and depends on M-phase-specific CDK-cyclin activity. Finally, a spindle checkpoint controls the activity of the APC/C and guards the metaphase–anaphase transition by assuring that all chromosomes are aligned on the equatorial plate and are attached to the mitotic spindle. In yeast and animals, several meiotic checkpoints have been identified that roughly correspond to these mitotic checkpoints. In contrast, meiotic checkpoints appear to be very differently setup in plants.
The pachytene checkpoint has been found to rely on many of the components of the mitotic DNA damage checkpoint and meiotic arrest is alleviated if mitotic DNA damage checkpoint components are inactivated. In particular, the pachytene checkpoint has been found to depend on Wee1-type kinases, which catalyze phosphorylation of highly conserved Thr and/or Tyr residues in the P-loop of Cdk1-type kinases and by that block their activity
The relaxed nature of the pachytene checkpoint in plants could at least be partially due to different mechanisms of how plants arrest the cell cycle after DNA damage. Although WEE1 homologues exist in plants and have for instance been isolated from maize, tomato, and Arabidopsis x WEE1 function appears to have undergone functional diversification since at least in Arabidopsis, wee1mutants neither have mitotic problems nor are impaired to arrest the cell cycle after DNA double-strand breaks. Also, dephospho-mutants in CDKA;1 that cannot be phosphorylated by WEE1 are viable and not hypersensitive to DNA-damaging drugs Consistently, recent observations suggested that instead of controlling cell cycle progression via CDKA;1, Arabidopsis WEE1 prevents premature cell differentiation after DNA damage in S phase in a yet unknown mechanism. (Wohlbold and Fisher 2009), (Lydall et al. 1996). (Sun et al. 1999; Sorrell et al. 2002; Gonzalez et al. 2004), (De Schutter et al. 2007; Cools et al. 2011) (Dissmeyer et al. 2009, 2010).
Marimuthu et al. (2011) described the construction of a spo11, rec8 osd11 triple mutant in Arabidopsis in the F1 of a cross between two natural accessions (i.e., a plant homozygous for the mutations, but heterozygous for all other alleles present between the two accessions). In this triple mutant, no recombination occurs; sister chromatids segregate at meiosis I and the second meiotic division is omitted. Consequently, these plants execute a mitosis-like meiotic cell division that produces viable diploid spores with a genotype identical to the parent. Since in Arabidopsis, haploid or diploid gametes can directly be grown into seeds and subsequently into plants; it was possible to grow offspring from this F1, which were identical to the mother plant, thereby effectively cloning the F1 through seeds (Since contemporary breeding relies heavily on heterozygous varieties that are preferred because of their higher yield this modulation of meiosis may show a way how to propagate heterozygote crops as clonal lines rather than creating them anew each year by crossing homozygous parental lines.
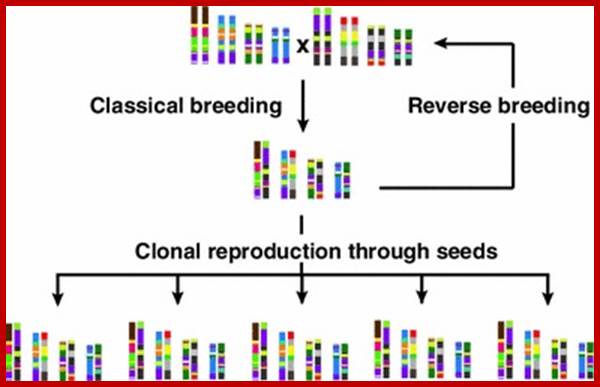
Relaxed meiotic checkpoints allow the development of new breeding approaches. Classical breeding refers to the classical method of constructing a hybrid by crossing two homozygous lines. Reverse breeding allows homozygous breeding lines to be constructed directly from a heterozygous parent essentially reversing classical breeding. Clonal reproduction through seeds allows the propagation of hybrids without homozygous intermediates. Please note that the given breeding schemes are simplified representations of these techniques. For further information please see Marimuthu et al. (2011), Wijnker et al. (2012) and Dirks et al. (2009) http://www.ncbi.nlm.nih.gov/ (Ravi and Chan 2010; Marimuthu et al. 2011), Marimuthu et al. 2011) . (Chen 2010),
Rice is a good model organism for exploring the molecular mechanisms of meiosis in higher plants. So far, 28 rice meiotic genes have been characterized byTos17 or T-DNA insertion site tagging, map-based cloning, as well as RNA interference silencing. So far, 28 rice meiotic genes have been characterized byTos17 or T-DNA insertion site tagging, map-based cloning, as well as RNA interference silence.
Cell cycle control must be modified at meiosis to allow two divisions to follow a single round of DNA replication, resulting in ploidy reduction. The mechanisms that ensure meiosis termination at the end of the second and not at the end of first division are poorly understood. We show here that Arabidopsis thaliana TDM1, which has been previously shown to be essential for meiotic termination, interacts directly with the Anaphase-Promoting Complex. Further, mutations in TDM1 in a conserved putative Cyclin-dependent Kinase (CDK) phosphorylation site (T16-P17) dominantly provoked premature meiosis termination after the first division, and the production of diploid spores and gametes. The CDKA1-CYCA1.2/TAM complex, which is required to prevent premature meiotic exit, phosphorylated TDM1 at T16 in vitro. Finally, while CYCA1;2/TAM was previously shown to be expressed only at meiosis I, TDM1 is present throughout meiosis. These data, together with epistasis analysis, lead us to propose that TDM1 is an APC/C component whose function is to ensure meiosis termination at the end of meiosis II, and whose activity is inhibited at meiosis I by CDKA;1-TAM-mediated phosphorylation to prevent premature meiotic exit. This provides a molecular mechanism for the differential decision of performing an additional round of division, or not, at the end of meiosis I and II, respectively.
@@
Starvation as external external chemical signal for diploid cells enter into meiotic cell cycle, in reproductive germ cells, but if rich medium is provided the cells return to mitotic phase. Meiotic cell cycle commitment to meiosis can be reversed, this holds good until the first meiotic division.
Mei2 RNA-binding protein works together with the noncoding meiRNA to initiate meiotic S-phase and also to promote subsequent events in early meiotic prophase I. Whereas meiotic mRNAs are eliminated during the mitotic cell cycle, Mei2 and meiRNA antagonize that elimination and induce meiotic entry. See reviews for additional information about this intriguing mechanism of mitosis/meiosis control.
In plants however, those germ cells, termed generically “sporocytes” in the two sexes or “megaspore mother cell” and “pollen mother cell” in females and males, respectively, typically enter into the meiotic cell cycle soon after their induction from the somatic hypodermal tissue. In Arabidopsis extrinsic signaling such as Leucine-rich receptor—like protein kinase controls germ line activation. Second intrinsic regulators control early meiotic prophase in both ovules and anthers. Switch1 and Ameiotic1 genes which encode homologous plant specific proteins get localized to the nucleus. Switch1 is required for histone modifications; Argonaute, germ cell specific controls early meiotic prophase and histone modifications in rice; these events requirement suggests the histone modifications are important for the cells to enter into meiotic phase. The first major regulatory transition occurs in late G1, when the Start of meiotic cycle is activated by Ime1 instead of Cln3/Cdk1 in mitosis.
The CHEK1 and CHEK2 play a central role in human and mouse meiosis events and their orthologs are also found in Saccharomyces cerevisiae, C.elegans, and Drosophila
The second phase is to make certain that the entry into metaphase1 that DNA replication is completed without any repairs left out. This even is regulated by the activation of M-Cdk in late prophase. The spindle assembly check point proteins check the attachment of MTs to kinetochores and APC cdc20 can initiate the process.
The second major transition occurs at the entry into metaphase I. The main purpose of this step is to make sure that DNA replication has completed without error so that spindle pole bodies can separate. The event is triggered by the activation of M-Cdk in late prophase I. Then the spindle assembly checkpoint proteins examine the attachment of microtubules at kinetochores and APCCdc20 can initiate the metaphase I. Chromosome separation in meiosis, homologous chromosomes separate in meiosis I and chromatids separation in meiosis II, requires special tension between homologous chromatids and non-homologous chromatids for distinguishing microtubule attachment and it relies on the programmed DNA double strand break (DSB) and repair in prophase I. Therefore meiotic recombination checkpoint can be a kind of DNA damage response at specific time spot. On the other hand, the meiotic recombination checkpoint also makes sure that meiotic recombination does happen in every pair of homologs.
Meiosis Stages;
Interphase: This stage is a preparatory stage for the subsequent events of karyokinesis. This, like mitosis, is also subdivided into G1, S & G2 stages. CHEK1 proteins play an important role in integrating DNA repair.
Meiosis is an important cell division in Diploid organisms, some may smaller and some may be bigger in size. Even plants during reproduction produce specific reproductive organs where they produce haploid cells which act as gametes.
In Angiosperm plants, male reproductive structure in the flower is stamen which produces Anthers; which in their chambers microspore mother cells undergoes meiosis and generates four haploid cells which develop into pollen grains. Each pollen grain consists of two haploid nuclei, when pollen grains fall on to the stigmatic surface they germinate and the pollen tube carries two nuclei. One fuses with the egg cell and the other fuses with secondary nucleus to produce triploid cell which develops into endosperm.
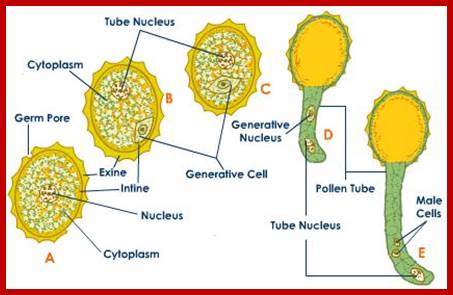
http://biology.tutorvista.com/
In angiosperms female sex organ in the flower is ovary in which ovules develop. Ovules as they mature produce megaspore mother cells which undergo meiotic cell division producing four cells of which 3 gradually degenerate. The lone single cell enlarges into embryo sac in which the single nucleus undergoes two more nuclear divisions to generate four nuclei, of which 3 organize into antipodal cells and three organize one Egg and two supporting synergids. Two nuclei fuse into secondary nucleus/polar nucleus. After fertilization one of the male nucleus fuse with the egg cell and the other fuses with secondary nucleus which develops into endosperm.
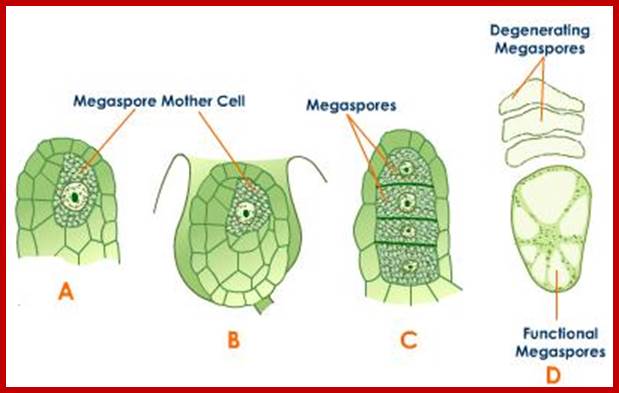
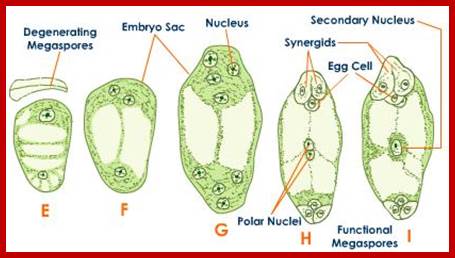
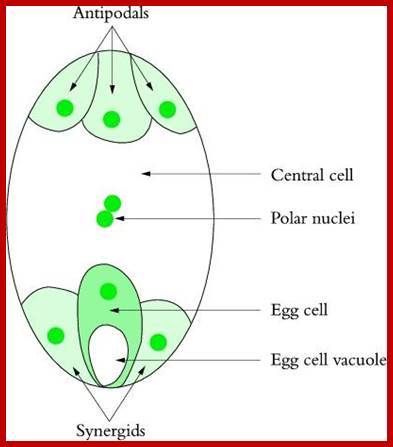
http://plantsinaction.science.uq.edu.au/
Meiotic transcriptomes in plants have been studied in Arabidopsis thaliana, rice (Oryza sativa), wheat (Triticum aestivum), petunia (Petunia hybrida), sunflower (Helianthus annuus), and maize (Zea mays). Studies in all organisms, but particularly in plants, indicate that a very large number of genes are expressed during meiosis, though relatively few of them seem to be required for the completion of meiosis.
In Arabidopsis meiocytes, approximately 20,000 genes are transcriptionally active; These genes constitute roughly 60% of the annotated genes in this species. In maize, about 50% of the 32,500 annotated genes are transcriptionally active during meiosis. But to date only about 90 genes are documented to act in Arabidopsis meiosis. About 50 of them are known to be essential for meiosis. ASY1 in Arabidopsis, which encodes a protein essential for homologous chromosome synapsis, is expressed in both reproductive and non-reproductive tissues, though the protein is only detected in meiocytes. On the other hand, expression of DMC1, a gene encoding a recombination protein that acts only in meiosis, is restricted to meiotic cells in anthers and carpels. For instance, RAD51 in Arabidopsis and maize, which encodes a protein facilitating DNA strand-exchange in meiotic recombination and somatic homologous recombination, is expressed predominantly in meiotic cells as well as developing embryos and seedlings.
In rice, microarray studies identified 2155 genes expressed at higher levels in meiotic anthers compared to seedlings. Many of these genes have not been previously linked to meiosis. A microarray study in petunia also found several novel meiotic genes; roughly 1,000 transposable elements, 32.5% of all transposable elements annotated genome-wide, are expressed in Arabidopsis meiocytes; Transposable elements belonging to the Copia, Gypsy, and SINE families exhibit the highest meiotic activity .
Biogenesis of these secondary siRNAs is triggered by microRNAs (miRNAs). The siRNAs are either 21 nt or 24 nt in size and produced in a phased manner, and hence named phased secondary siRNAs (phasiRNAs;). In addition to these small RNAs, several miRNAs known to be involved in transcriptional gene silencing have been detected in the meiosis RNA-seq studies of Arabidopsis Mi et al., 2008; Yang et al., 2011. Song et al., 2011, Wang et al., 2005. http://journal.frontiersin.org/;
Involvement of small RNAs in meiosis has also been documented outside of plants. PIWI proteins are small-RNA-binding proteins in the germline of metazoans that belong to the family of Argonaute (AGO) proteins. PiRNAs, small RNAs associated with PIWI, serve to silence transposons in the mouse male germline.
Little is known about what specifically phasiRNAs and miRNAs might do in meiosis in plants, and functional studies are needed in this area. Many transposable elements are also expressed during meiosis. Meiotic Recombination in Plants; http://phys.org/
Chromatin undergoes drastic structural and spatial reorganization in early meiotic prophase I This reorganization includes changes in histone modification patterns, chromosome condensation, and repositioning within the nucleus. Histone hyperacetylation was found to be required for proper recombination and chromosome segregation in Arabidopsis .
Though not much is understood about DNA methylation patterns in plant meiosis, there is evidence indicating that microspores in Arabidopsis exhibit altered DNA methylation patterns compared to other reproductive tissues (Calarco et al., 2012). This evidence could suggest that during meiosis, there are changes in DNA methylation patterns, though more research is needed to test this hypothesis. Many RNAi and LNC RNA are expressed and their genes is upregulated during meiosis. Genes known to participate in the RNAi pathway have been found to be up-regulated in meiosis, The most promising example is a novel class of secondary short interfering RNAs (siRNAs) that have been recently discovered in cereals and are preferentially expressed in the stamen . The siRNAs are either 21 nt or 24 nt in size and produced in a phased manner, and hence named phased secondary siRNAs (phasiRNA). In addition to these small RNAs, several miRNAs known to be involved in transcriptional gene silencing have been detected in the meiosis RNA-seq studies of Arabidopsis.
In contrast to small RNAs which are 21–24 nt in length, long non-coding RNAs (lncRNAs) are generally >200 bp in length and polyadenylated). They form secondary structures which allow them to interact with other nucleic acid molecules to function in activating and repressing genes, as well as in epigenetic modification of chromatin. Since the discovery of the first lncRNA, Xist, in mammals and its role in X-chromosome inactivation, thousands of lncRNAs have been identified. While a number of lncRNAs have indeed been found to play roles in regulating gene expression, the functions of the vast majority of them are unknown. A transcriptome study in mouse indicated that roughly 8000 lncRNAs were expressed in spermatids and spermatocytes. Presence of these lncRNAs is not a result of ectopic read-through transcription but likely a controlled activity ((Nam and Bartel, 2012; Soumillon et al., 2013;Margolin et al., 2014, Lee, 2012. (Dawe et al., 1994; Zickler and Kleckner, 1998; MacQueen and Villeneuve, 2001; Kimmins and Sassone-Corsi, 2005; Yang et al., 2006; Sheehan and Pawlowski, 2009). http://www.ncbi.nlm.nih.gov/
Human male and female Ovary and Testes;http://www.biology.iupui.edu/
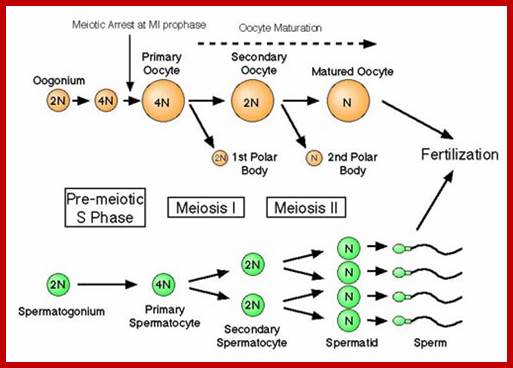
In animal systems, for example, let us take humans- Male sex organ is testis. In testis, as it matures with human growth, spermatogenesis is induced. This leads to production of haploid motile sperms.

The SRY of Male sex chromosome direct male development in mammals by inducing the somatic cells of gonadal ridge to develop into testis; Each time a spermatogonium divides it may either undergo mitosis, to maintain the number of spermatogonia, or spermatogenesis. Spermatogenesis begins with meiosis 1 and to produce two cells which undergo one more meiotic division to produce four sperm cells. Male germ cells interact with seroli cells via nodal signaling in human seminiferous tubules; serotoli clls also produce growth factors of human serotoli cells; this leads to spermatogenesis; ttp://www.ajandrology.com/ http://slideplayer.com/
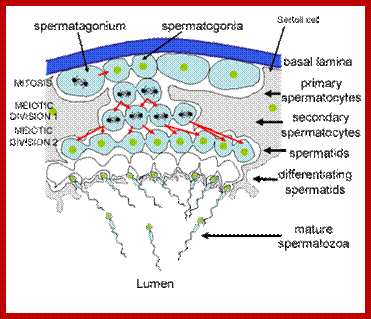
http://www.histology.leeds.ac.uk/
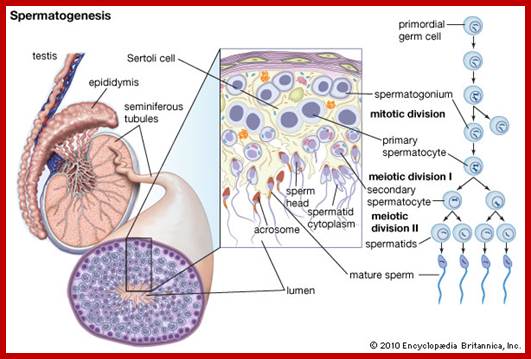
https://www.britannica.com/science/

http://www.biology.lifeeasy.org
Spermiogenesis- during which the following
transformations takes place. 1.Axonemal structure, 2. Golgi complex, 3.
Acrosomal vescicle, 4. Pair of centrioles, 5. Mitochondria, 6. Nucleus, 7.
Flagella primordium, 8. Microtubules, 9. Sperm cell tails,10. Acrosomal cap.
Spermiogenesis: The differentiation of the spermatids into sperm cells is called spermiogenesis. It corresponds to the final part of spermatogenesis and comprises the following individual processes that partially proceed at the same time: Differentiation of spermatids to sperm cells is called spermiogenesis.
- Nuclear condensation: thickening and reduction of the nuclear size, condensation of the nuclear contents into the smallest space. The nucleus becomes smaller, denser and takes on a characteristic, flattened form. Seen from above, the nucleus is oval and, from the narrow side, is pear-shaped. The acrosome lies over the tip. Nucleus and acrosome form the sperm cell's head that is bound to the mid-piece by a short neck.
- Acrosome formation: Forming a cap (acrosome) containing enzymes that play an important role in the penetration through the pellucid zone of the oocyte. The Golgi complex engender the vesicles, which then merge into a larger formation that settles close to the cell nucleus and finally inverts itself like a cap over the largest part of the nucleus. The acrosome corresponds functionally to a lysosome and thus contains lysosomal enzymes (hyaluronidase among others).
- Flagellum formation: generation of the sperm cell tail. The future axonemal structure grows out of one centriole (distal). This consists of a bundle of nine peripheral double microtubules and two single ones in the center. During its development, through the rotation of the nucleus and acrosomal vesicle, the flagellum primordium comes to lie on the opposite side of the acrosome.
- Cytoplasm reduction: elimination of all unnecessary cytoplasm.

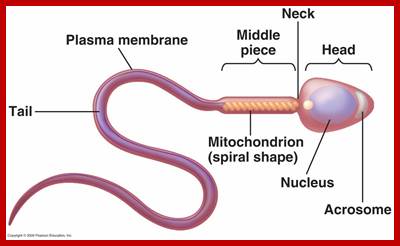
http://www.biology.lifeeasy.org/
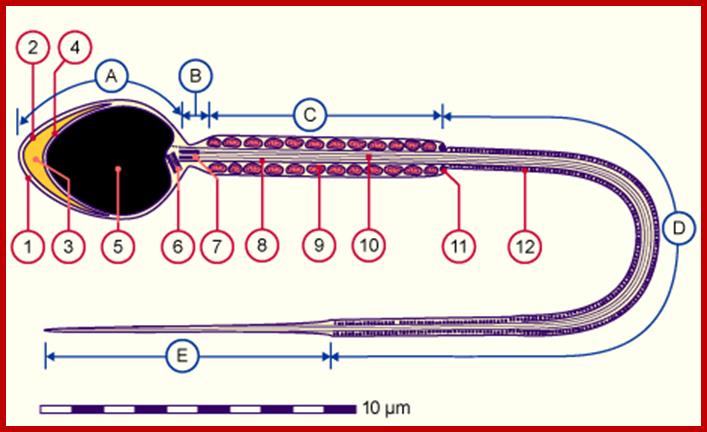
1.Plasma membrane, 2.Outer acrosomal membrane, 3.Acrosome, 4.Inner acrosomal membrane, 5.Nucleus, 6.Proximal centriole, 7.Rest of the distal centriole, 8.Thick outer longitudinal fibers, 9.Mitochondrion, 10.Axoneme, 11.Anulus, 12.Ring fibers; http://www.biology.lifeeasy.org/95
Head: It is spherical in shape consisting of large nucleus and a dome shaped acrosome present on the nucleus. Function: Nucleus contain genetic information and half number of chromosomes. The acrosome releases a hyaluronidase enzyme which destroys the hyaluronic acid of the ovum and enters into the ovum.
Neck: It contains centrioles which are proximal centriole and distal centriole. Function: Distal centriole gives rise to axial filament of the sperm which runs up to the end of the tail.
Middle piece: It is tubular structure in which mitochondria are spirally arranged. Function: Middle piece is called power house of sperm because it gives energy to the sperm to swim in the female genital tract.
Tail: It arises from middle piece and it is the end part of the sperm. It contains axial filaments. Function: Tail helps the sperm to swim in the female genital tract. It is the main part of sperm to move.
Ion fluxes during mammalian sperm hyper activation and capacitation, where both processes operate during sperm transition through female tract and contain intercrossing pathways. Albumin present in the female tract removes cholesterol from the sperm plasma membrane modifying several membrane properties; it may also directly activate Cat-Sper channels, increasing [Ca2+] ions. External Ca2+ ions activate Na+/HCO3− (NBC) cotransporters (possibly activated by external Ca2+ this increases HCO3− levels activating a soluble adenylyl cyclase (sAC) and producing cAMP. Then cAMP activates the sperm Na+/H+ exchanger (sNHE) and together with the activation of the proton channel (HV) (by Zn2+ removal) would raise pH. Another cellular parameter activates CatSper and SLO3 channels.
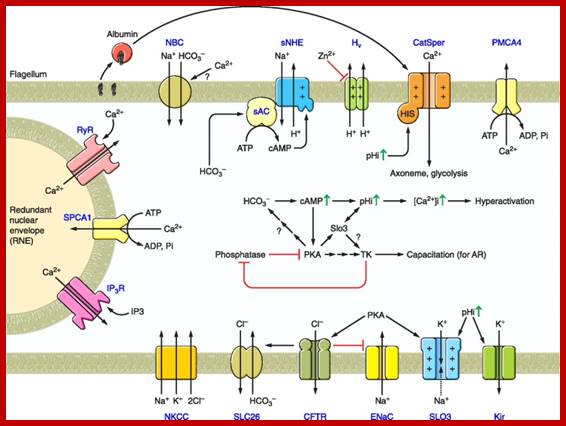
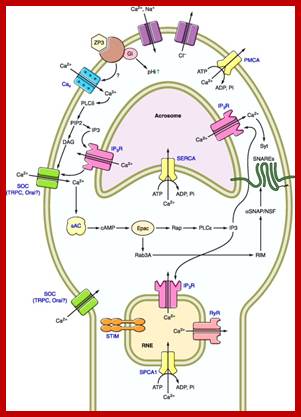
http://physrev.physiology.org/
The overall Ca2+ increase may influence glycolysis and the axoneme activity promoting hyper activation of motility. Several Ca2+ mobilizing pumps (PMCA4, SPCA1) and channels [IP3 receptor (IP3R), ryanodine receptor (RyR)] may also participate during Ca2+ signalling. Concomitantly, the increase in cAMP levels activates PKA, which after several unknown steps stimulates tyrosine kinases to produce the tyrosine phosphorylation associated with capacitation. Possible connections between hyper activation and capacitation signalling cascades are indicated. In some species, capacitation is accompanied by membrane hyperpolarization; the channels and transporters involved during this process are head, neck, midpiece and principal piece.
Mammalian spermatogenesis is the representative for most animals. In human males, spermatogenesis begins at puberty in seminiferous tubules in the testicles and goes on continuously. Spermatogonia are immature germ cells. They proliferate continuously by mitotic divisions around the outer edge of the seminiferous tubules, next to the basal lamina. Some of these cells stop proliferation and differentiate into primary spermatocytes. After they proceed through the first meiotic division, two secondary spermatocytes are produced. The two secondary spermatocytes undergo the second meiotic division to form four haploid spermatids. These spermatids differentiate morphologically into sperm by nuclear condensation, ejection of the cytoplasm and formation of the acrosome and flagellum. Spermatogenesis- several generation of mitosis –growth- then Meiosis cycles- to generate sperms.
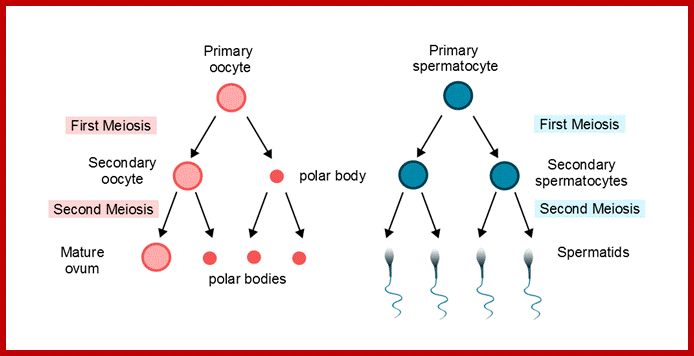
Diagram of oogenesis and spermatogenesis
with regard to polar bodies
Diagram courtesy of Sarah Lee; http://www.histocutup.co.uk/Gynaecology

http://www.biology.iupui.edu/
In human females, oogenesis begins when the female embryo is still in the womb. Meiosis goes through diplonema of Meiosis 1 and further progress is arrested till reaching puberty in females; then with time small clutches of these arrested oocytes will proceed up to metaphase II and await fertilization.
One oocyte (2n) produces one egg; during first meiotic metaphase plate in dividing germ cell, instead of placing in the middle, it is tucked in the margin of the dividing cell. This results in equal distribution of genetic material, but leads to unequal distribution of cytoplasm and its associated organelles as cytokinesis takes place. First division results in one large cell and another small cell called polar body. This cell proceeds to second division, where large cell divides to produce one small cell called polar body cell and the large cell is called the egg. The egg is large with most of the original cytoplasm including all organelles especially mitochondria. In this mode oogonia produces 5-7 million but just 5000 or less remain viable. Once reaching puberty female generate one egg every 23-28 days till the female reaches 57- 60 years when production of eggs stops- the stage is called menopause. In males, sperm production continues as long as he can exercise for sex.
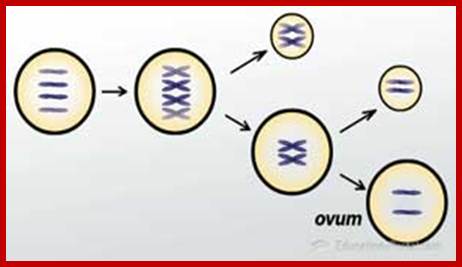
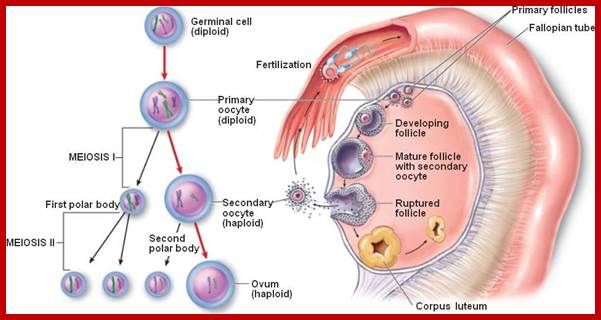
http://biology4isc.weebly.com/
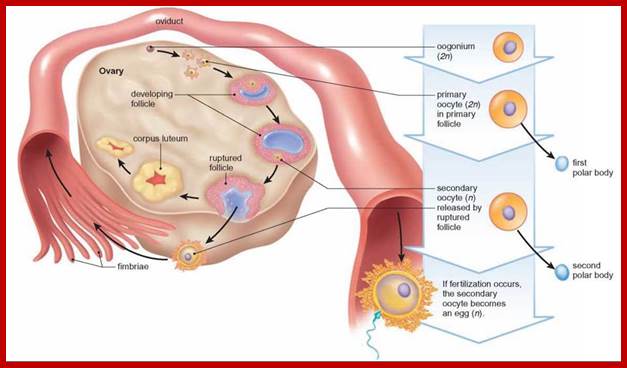
After puberty the ovary goes through a regular monthly cycle that involves growth of follicles, ovulation, and the development of a corpus luteum;
http://schoolbag.info/
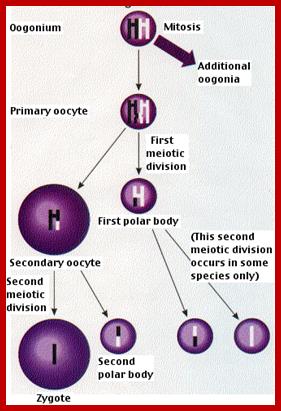
http://www.biology-questions-and-answers.com/-Gametogenesis
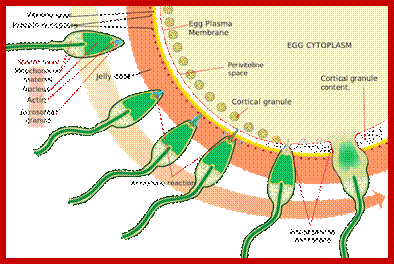
https://en.wikipedia.org/wiki/
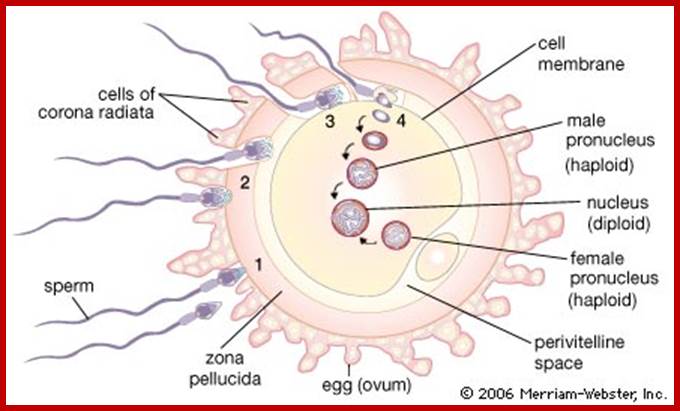
https://www.britannica.com
Fertilization in humans, sperm fusion with the egg takes place in the Ampulla of the fallopian tube. The sperm undergoes capacitation in the female reproductive sac. The sperm encountering the egg cell surface, sperm plasma membrane fuses with egg plasma membrane; sperms acrosome has all the enzymes for penetration via digesting many of the obstacle components on the egg surface. During the entry of the sperm all of its mitochondria are released out, even if some are remained adhered, they are also destroyed by the egg cytoplasmic components.
During fertilization, a sperm must first fuse with the plasma membrane and then penetrate the female egg in order to fertilize it. Fusing to the egg usually causes little problem, whereas penetrating through the egg's hard shell can present more of a problem to the sperm. Therefore, sperm cells go through a process known as the acrosome reaction which is the reaction that occurs in the acrosome of the sperm as it approaches the egg. The acrosome is a cap-like structure over the anterior half of the sperm's head.
As the sperm approaches the zona pellucida of the egg, which is necessary for initiating the acrosome reaction, the membrane surrounding the acrosome fuses with the plasma membrane of the oocyte, exposing the contents of the acrosome. The contents include surface antigens and numerous enzymes which are responsible for breaking through the egg's tough coating and allowing fertilization to occur.
The sperm may trigger egg activation via the interaction between a sperm protein and an egg surface receptor. Izumo is the sperm cell signal that will trigger the egg receptor Juno. This receptor is activated by the sperm binding and a possible signaling pathway could be the activation of a tyrosine kinase which then activates phospholipase C (PLC). The inositol signaling system has been implicated as the pathway involved with egg activation. IP3 and DAG are produced from the cleavage of PIP2 by phospholipase C. However, another hypothesis is that a soluble 'sperm factor' diffuses from the sperm into the egg cytosol upon sperm-oocyte fusion. The results of this interaction could activate a signal transduction pathway that uses second messengers. A novel PLC isoform, PLC-ζ, may be the equivalent of the mammalian sperm factor. A 2002 study demonstrated that mammalian sperm contain PLC zeta which can start the signaling cascade.
It is at this time of fertilization, the egg cell progresses to meiosis II; at this point the large secondary nucleus undergoes final division produce a large egg cell and smaller polar body. Then the sperm nucleus fuses with egg nucleus. The most important feature of fertilization is that all male sperm carrying mitochondria does not contribute any of its male mitochondria to egg. Thus whatever mitochondria found in the born progeny are of mitochondria derived from mother not male donated sperm bound mitochondria. This mitochondrial inheritance vis a females started in African few mothers where the humans originated and developed into a population. This mitochondrial DNA can be used to identify the individual of descent in the population. During fertilization, a sperm must first fuse with the plasma membrane and then penetrate the female egg in order to fertilize it. Fusing to the egg usually causes little problem, whereas penetrating through the egg's hard shell can present more of a problem to the sperm. Therefore, sperm cells go through a process known as the acrosome reaction which is the reaction that occurs in the acrosome of the sperm as it approaches the egg. The acrosome is a cap-like structure over the anterior half of the sperm's head.
Four hours after fusion of sperm and ovum, DNA synthesis begins. Male and female pronuclei move to the Centre of the egg and membranes break down. Male protamines are replaced with histones and the male DNA is demethylated. Chromosomes then orientate on the metaphase spindle for mitosis. This combination of the two genomes is called Syngamy.
It is important to note- Only if fertilization occurs meiosis II will be completed. Entry of the sperm restarts the cell cycle breaking down MPF (M-phase promoting factor) and turning on the anaphase-promoting complex (APC). Completion of meiosis II converts the secondary oocyte into a fertilized egg or zygote
http://www.aboutthemcat.org/

http://www.aboutthemcat.org/
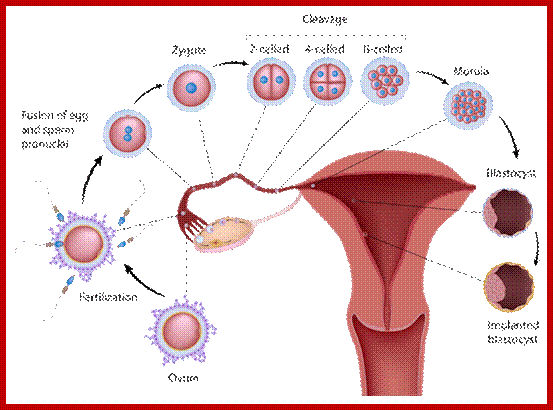
Internal Checkpoints during the Cell Cycle;
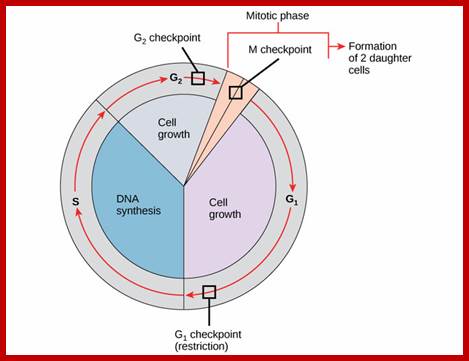
Boundless Biology; http://jcs.biologists.org/
Checkpoints are regulatory mechanisms that block cell cycle progression when key cellular processes are defective or chromosomes are damaged. During meiosis, genetic recombination between homologous chromosomes is essential for proper chromosome segregation at the first meiotic division. In response to incomplete recombination, the pachytene checkpoint (also known as the meiotic recombination checkpoint) arrests or delays meiotic cell cycle progression, thus preventing the formation of defective gametes.
Internal Checkpoints during the Cell Cycle;
Here, we describe a role for a meiosis-specific kinase, Mek1, in the meiotic recombination checkpoint in fission yeast. Mek1 belongs to the Cds1/Rad53/Chk2 family of kinases containing fork head-associated domains, which participate in a number of checkpoint responses from yeast to mammals.
The cell cycle is controlled at three phases. The integrity of the DNA is assessed at the G1 checkpoint. Proper chromosome duplication is assessed at the G2 checkpoint. Attachment of each kinetochore to a spindle fiber is assessed at the M checkpoint. During the meiotic cell cycle, a surveillance mechanism called the “pachytene checkpoint” ensures proper chromosome segregation by preventing meiotic progression when recombination and chromosome synapsis are defective. The silencing protein Dot1 (also known as Pch1) is required for checkpoint-mediated pachytene arrest of thezip1 and dmc1 mutants of Saccharomyces cerevisiae. In the absence of DOT1,(Pch1) the zip1and dmc1 mutants inappropriately progress through meiosis, generating unviable meiotic products. http://www.ncbi.nlm.nih.gov.
Other components of the pachytene checkpoint include the nucleolar protein Pch2 and the heterochromatin component Sir2. In dot1, disruption of the checkpoint correlates with the loss of concentration of Pch2 and Sir2 in the nucleolus.
In Saccharomyces cerevisiae, several components of the DNA damage checkpoint have been identified. Rad9 and the Rad24 group of proteins (Rad24, Rad17, Mec3, and Ddc1) are thought to be involved in sensing damage and/or generating a signal in response to damage.
The same checkpoint controls that arrest progression of the mitotic cell cycle in response to DNA damage, blocks in replication, or defects in spindle integrity also function in meiosis; In addition, meiotic cells possess a surveillance mechanism called the “pachytene checkpoint” that monitors events specific to meiosis, such as recombination and chromosome synapsis (i.e., SC formation), that are critical for proper meiotic chromosome segregation, in Saccharomyces cerevisiae zip1 mutant, which exhibits defects in synaptonemal complex formation and meiotic recombination, triggers a checkpoint that causes cells to arrest at the pachytene stage of meiotic prophase. These results suggest that meiotic chromosomal proteins function in the signaling of meiotic prophase defects and that the correct stoichiometry of Red1, Mek1, and Hop1 is needed to achieve checkpoint-mediated cell cycle arrest at pachytene.
However, in cells in which the function of the DNA-replication-checkpoint is the DSBs in the HU-treated mutant cells occurred at normal sites and were associated with recombination. In addition, Cdc2p is apparently not involved in this process. We propose that the sequence of meiotic S phase and initiation of recombination is coordinated by DNA-replication-checkpoint proteins.
The spindle assembly checkpoint prevents chromosome mis-segregation during both mitosis and meiosis. In meiosis kinetochore microtubule attachment is not clear. In meiosis a weakened check point could contribute age-related aneuploidy found in humans. In mitosis this failure may end up in tumor progression if tumours have initiated.
Meiotic check point protein kinases CHEK1 and Check2 play central role in meiosis of human and mouse and their orthologues in Saccharomyces cerevisiae; Caenorhabditis elegans, Schizosaccharomyces pombe and Drosophila has been reviewed. During meiotic recombination in human and mouse, CHEK1 protein kinase is important for integrating DNA damage repair with cell cycle arrest. CHEK1 is expressed in the testes; and it is associated with meiotic synaptonemal complexes during zygonema and pachynema stages. CHEK1 likely acts as an integrator for ATM and ATR signals and in monitoring meiotic recombination. In mouse oocytes CHEK1 appears to be indispensable for prophase I arrest and to function at the G2/M checkpoint. MacQueen and Hochwagen; Subramanian and Hochwagen
CHEK2 regulates cell cycle progression and spindle assembly during mouse oocyte maturation and early embryo development. Although CHEK2 is a downstream effector of the ATM kinase that responds primarily to double-strand breaks it can also be activated by ATR (ataxia-telangiectasia and Rad3 related) kinase that responds primarily to single-strand breaks. In mouse, CHEK2 is essential for DNA damage surveillance in female meiosis. The response of oocytes to DNA double-strand break damage involves a pathway hierarchy in which ATR kinase signals to CHEK2 which then activates p53 and p63 proteins.
The Saccharomyces cerevisiae zip1 mutant, which exhibits defects in synaptonemal complex formation and meiotic recombination, triggers a checkpoint that causes cells to arrest at the pachytene stage of meiotic prophase. Imbalance of meiotic chromosomal proteins inactivates the pachytene checkpoint. Overproduction of Red1 or Mek1 specifically promotes sporulation of mutants that normally undergo checkpoint-mediated arrest at pachytene. Thezip1, zip2, dmc1, and hop2 mutants all exhibit defects in both recombination and synapsis; however, the molecular signal that triggers arrest in these strains remains unknown. http://mcb.asm.org/;http://www.pnas.org/;http://mcb.asm.org; http://onlinelibrary.wiley.com/
A model of functions of the checkpoint clamp and clamp loader in crossover formation: Four possible pathways of meiotic recombination are shown. Rad51 and Dmc1 cooperate in pathways that display inter-homolog bias (three leftmost pathways). Together with Rad51–Dmc1, the 9‐1‐1 clamp and Mec1 suppress inter-sister and ectopic recombination (right pathway). ZMM proteins specifically promote the ‘crossover‐only’ pathway (third pathway from the left) by processing inter-homolog joint molecules – this depends on Hop1–Red1–Mek1. The 9‐1‐1 clamp, but not Mec1, facilitates ZMM function on the crossover‐only pathway. ZMM proteins might also promote inter-sister joint molecule formation. Non‐crossover formation is independent of ZMM proteins (the leftmost pathway). Even in the absence of ZMM function, inter-homolog joint molecules are formed (the second pathway from the left). In this pathway, joint molecules are resolved into either crossover or NCO. http://jcs.biologists.org/
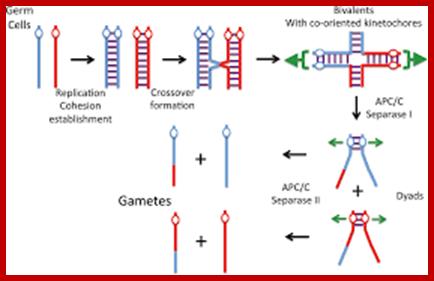
http://onlinelibrary.wiley.com/
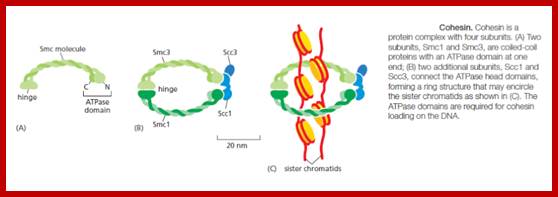
http://onlinelibrary.wiley.com/
Cohesin complex consists of SCC2/SCC4 is loaded on to meiotic leptotene chromosomes so that homologous chromosomes are paired properly in Zygotene.
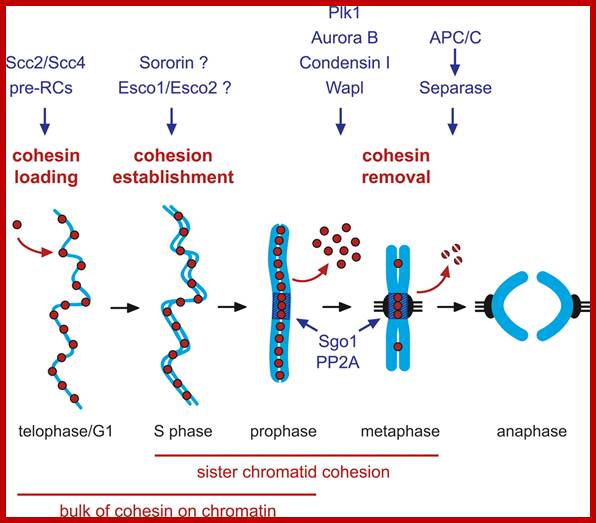
http://reasonandscience.heavenforum.org/
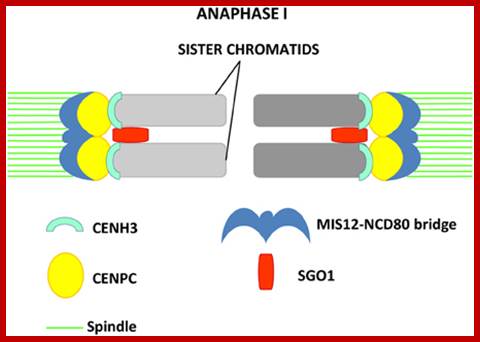
Model proposed by Li and Dawe (2009) for reductional segregation I Meiosis 1; sister chromatids are fused in meiosis 1; MIS12-NCD80 form the bridge; the inner Kinetochore proteins CENPC and CENH3 are visialized as two distinct signals; http://journal.frontiersin.org/
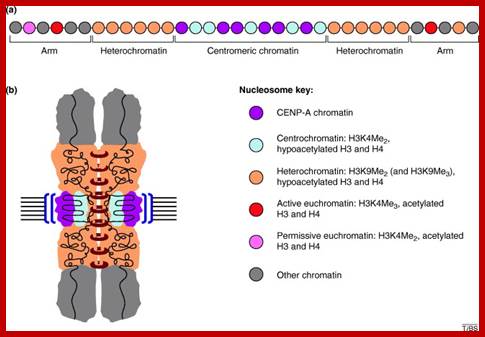
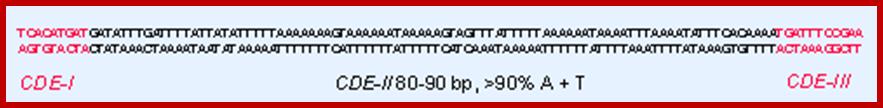
Post-translational modifications of core histones, particularly in the N terminus, seem to define different chromatin states in the genome. For instance, actively transcribed regions of euchromatin have a completely different set of ‘marks’ compared with silent heterochromatin. Recent analyses surprisingly demonstrate that, in addition to containing the histone H3 variant CENP-A, the centromeric chromatin that underlies the kinetochore bears a distinct combination of histone H3 modifications. Three conserved regions can be identified by the sequence homologies between yeast CEN elements; CDE 1 from the left 9bp, CDE II middle 90% rich in A+T, reminiscent of Satellite DNA and CDEIII 11bp conserved right; CEN region is 10bp long and it is nuclease resistant; http://www.cell.com/; C.David Allis, et albrinkley@bcm.tmc.edu; http://genes.atspace.org/
1.
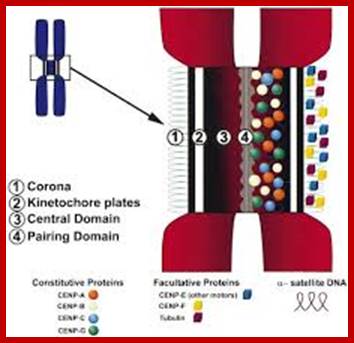
The mammalian centromere: structural domains and the attenuation of chromatin modeling; Aron A,Van Hoosser and Michael A.Mancin.,
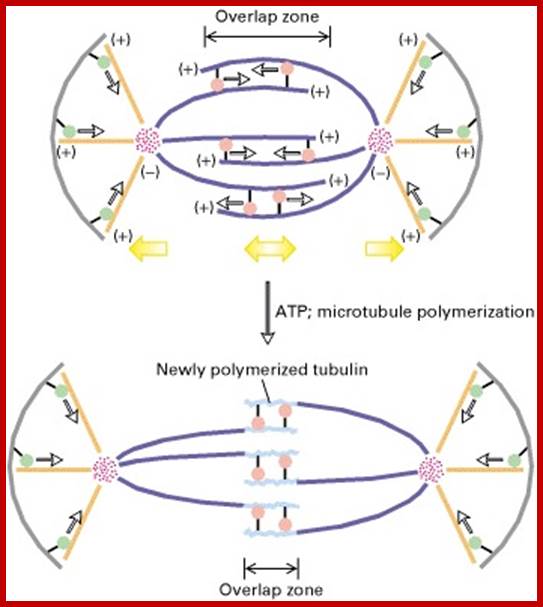
Adapted from H. Masuda and W. Z. Cande, 1987, Cell 49:193.
Model of spindle elongation and movement during anaphase B. Tubulin (light purple) adds to the (+) ends of all polar microtubules, lengthening these fibers.
Simultaneously (+) end – directed KRPs (pink) bind to the polar microtubules in the overlap region; each KRP, bound to a microtubule in one half-spindle, “walks” along a microtubule in the other half-spindle, toward its (+) end, utilizing the energy from ATP hydrolysis. In cells that assemble an aster, a (−) end – directed motor protein (light green) located in the cortex of the plasma membrane pulls on the astral microtubules, which also moves the poles farther apart.
Meiotic check point protein kinases CHEK1 and CHEK2:
The central role in meiosis of human and mouse CHEK1 and CHEK2 and their orthologs in Saccharomyces cerevisiae, Caenorhabditis elegans, Schizosaccharomyces pombe and Drosophila has been reviewed During meiotic recombination in human and mouse, CHEK1 protein kinase is important for integrating DNA damage repair with cell cycle arrest.
Meiotic check point protein kinases CHEK1 and CHEK2 play central role in meiosis of human and mouse and their orthologues in Saccharomyces cerevisiae; Caenorhabditis elegans, Schizosaccharomyces pombe and Drosophila has been reviewed.
During meiotic recombination in human and mouse, CHEK1 protein kinase is important for integrating DNA damage repair with cell cycle arrest. CHEK1 is expressed in the testes; and it is associated with meiotic synaptonemal complexes during zygonema and pachynema stages. CHEK1 likely acts as an integrator for ATM and ATR signals and in monitoring meiotic recombination. In mouse oocytes CHEK1 appears to be indispensable for prophase I arrest and to function at the G2/M checkpoint. MacQueen and Hochwagen; Subramanian and Hochwagen
CHEK2 regulates cell cycle progression and spindle assembly during mouse oocyte maturation and early embryo development. Although CHEK2 is a downstream effector of the ATM kinase that responds primarily to double-strand breaks it can also be activated by ATR (ataxia-telangiectasia and Rad3 related) kinase that responds primarily to single-strand breaks. In mouse, CHEK2 is essential for DNA damage surveillance in female meiosis. The response of oocytes to DNA double-strand break damage involves a pathway hierarchy in which ATR kinase signals to CHEK2 which then activates p53 and p63 proteins.
The Saccharomyces cerevisiae zip1 mutant, which exhibits defects in synaptonemal complex formation and meiotic recombination, triggers a checkpoint that causes cells to arrest at the pachytene stage of meiotic prophase. Imbalance of meiotic chromosomal proteins inactivates the pachytene checkpoint. Overproduction of Red1 or Mek1 specifically promotes sporulation of mutants that normally undergo checkpoint-mediated arrest at pachytene. Thezip1, zip2, dmc1, and hop2 mutants all exhibit defects in both recombination and synapsis; however, the molecular signal that triggers arrest in these strains remains unknown. http://mcb.asm.org/;http://www.pnas.org/;http://mcb.asm.org; http://onlinelibrary.wiley.com/
CHEK1 is expressed in the testes and associates with meiotic synaptonemal complexes during the zygonema and pachynema stages. CHEK1 likely acts as an integrator for ATM and ATR signals and in monitoring meiotic recombination. In mouse oocytes CHEK1 appears to be indispensable for prophase I arrest and to function at the G2/M checkpoint.
CHEK2 regulates cell cycle progression and spindle assembly during mouse oocyte maturation and early embryodevelopment. by MacQueen and Hochwagen; Subramanian and Hochwagen; http://journals.plos.org/plosgenetics/
CHEK1 and Check II function at prophase1 checking ds DNA break and repair and function at G2/M point. Although CHEK2 is a downstream effector of the ATM kinase that responds primarily to double-strand breaks it can also be activated by ATR (ataxia-telangiectasia and Rad3 related) kinase that responds primarily to single-strand breaks. In mouse, CHEK2 is essential for DNA damage surveillance in female meiosis. The response of oocytes to DNA double-strand break damage involves a pathway hierarchy in which ATR kinase signals to CHEK2 which then activates p53and p63 proteins.
Meiotic chromosomal recombination is checked by meiosis check points. These DNA breaks must be repaired before metaphase I. and these DSBs must be repaired before metaphase I. The cell monitor these DSBs via ATM pathway, in which Cdc25 is suppressed when DSB lesion is detected. This pathway is the same as classical DNA damage response and is the part we know the best in meiotic recombination checkpoint.
A surveillance mechanism called the “pachytene checkpoint” ensures proper chromosome segregation by preventing meiotic progression when recombination and chromosome synapsis are defective. DSBs occur naturally in the germline during meiosis but whether RAD9A participates in repairing such breaks is not known. Defects in recombination and/or chromosome synapsis activate the pachytene checkpoint, which delays meiotic cell cycle progression to avoid aberrant chromosome segregation and formation of defective gametes.
Jcs bioloist .org
Role of small RNAs and lincRNAs in Meiosis:
Biogenesis of these secondary siRNAs is triggered by microRNAs (miRNAs). The siRNAs are either 21 nt or 24 nt in size and produced in a phased manner, and hence named phased secondary siRNAs (phasiRNAs). In addition to these small RNAs, several miRNAs known to be involved in transcriptional gene silencing have been detected in the meiosis RNA-seq studies of Arabidopsis. They are also found genetic and epigenetic silencing pathways. RNAi-mediated DNA damage repair occurs in plants, which goes in line with previous studies in the fungus Neurospora crassa and S. pombe, in which rDNA-derived small RNAs and centromeric small RNAs, respectively, are induced upon DNA damage.
Involvement of small RNAs in meiosis has also been documented outside of plants. PIWI proteins are small-RNA-binding proteins in the germ line of metazoans that belong to the family of Argonaute (AGO) proteins. PiRNAs, small RNAs associated with PIWI, serve to silence transposons in the mouse male germ line
Little is known about what specifically phasiRNAs and miRNAs might do in meiosis in plants, and functional studies are needed in this area. Song et al., 2011), (Mi et al., 2008; Yang et al., 2011).
Meiotic non-coding RNA in Plants; http://phys.org/
Modes of action of non-coding RNAs in transcriptional and other regulations of genes in meiosis; (A) Long non-coding RNAs can participate in gene activation and suppression. (B) Long non-coding RNAs can act as a guide for chromatin modifications.
In contrast to small RNAs which are 21–24 ntds in length, long non-coding RNAs (lncRNAs) are generally >200 ntds long and polyadenylated (Nam and Bartel, 2012). They form secondary structures which allow them to interact with other nucleic acid molecules to function in activating and repressing genes, as well as in epigenetic modification of chromatin.
Since the discovery of the first lncRNA, Xist, in mammals and its role in X-chromosome inactivation, thousands of lncRNAs have been identified. While a number of lncRNAs have indeed been found to play roles in regulating gene expression, the functions of the vast majority of them are unknown (Lee, 2012). Transcriptome study in mouse indicated that roughly 8000 lncRNAs were expressed in spermatids and spermatocytes. Presence of these lncRNAs is not a result of ectopic read-through transcription but likely a controlled activity (Soumillon et al., 2013;Margolin et al., 2014). http://www.ncbi.nlm.nih.gov/
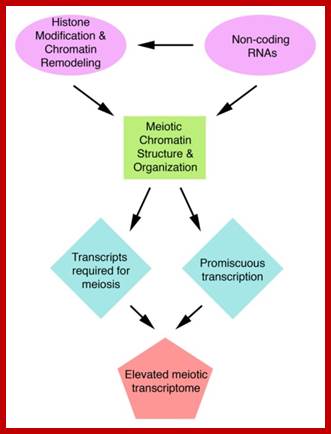
A diagram showing potential mechanisms that could affect the meiotic Transcriptome; http://www.ncbi.nlm.nih.gov/core
In some germ line cells some AGO proteins are expressed involved in hromosomal segregation nd cell fate specification. Small RNAs with AGO proteins as unlike miRNAs, secondary siRNA duplexes have good complementarity between guide and passenger strands have perfect complementarity between guide and passenger strands, so that sorting into different AGOs must rely exclusively on their 5′ terminal nucleotides. It has been observed recently, 24 ntds long phasiRNAs accumulate during entire meiosis and found in large quantities in pollen cells. Another group of 21-nucleotide phasiRNAs (phased small –interfering RNAs) accumulates in MEIOSIS ARRESTED AT LEPTOTENE 1 (MEL1) in rice, while its closest ortholog in maize is AGO5c, which seems to be expressed along with 21-nucleotide phasiRNAs in anthers. AGO5 and MEL1 are also expressed in ovules in involved in DNA methylation of heterochromatin. Diversification of small-RNA-directed silencing pathways through the expansion of RNA-dependent RNA polymerases, DICER proteins and ARGONAUTE proteins is well documented. Even PiRNA (pI-interacting RNAs) are also involved in meiosis.
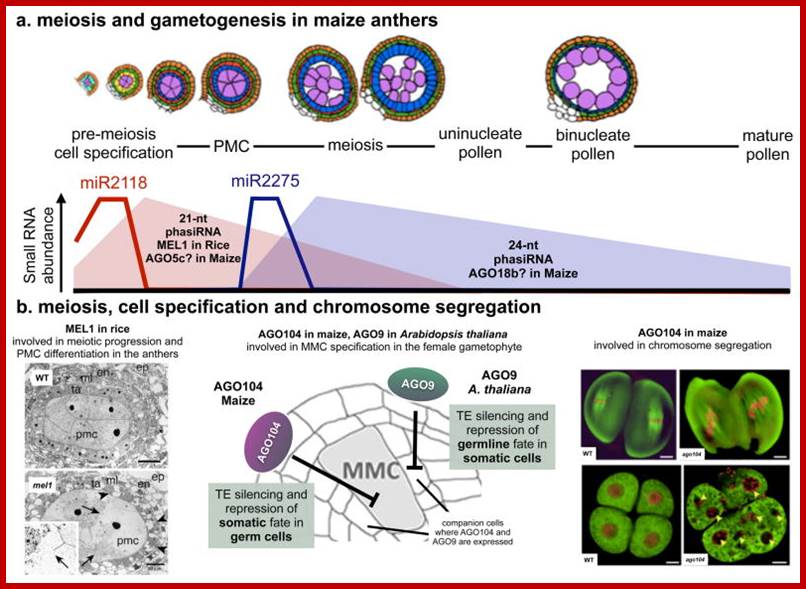
Small RNA functions in meiosis and cell fate specification;
a. In grass anthers, two distinct small RNA classes are produced from non-coding PHAS transcripts: 21-nucleotide (nt) phasiRNAs are produced upon cleavage of PHAS transcripts by miR2118, whereas miR2275 triggers 24-nucleotide phasiRNA biogenesis from a different subset of PHAS loci (reviewed in). The spatiotemporal dynamics of phasiRNA biogenesis was recently described throughout anther development in maize72, showing a distinct and mostly non-overlapping accumulation patterns for both phasiRNA classes, which nicely coincides with the expression of their respective miRNA triggers. 21-nucleotide phasiRNAs are essentially pre-meiotic, whereas 24-nucleotide phasiRNAs peak during meiosis and decrease during pollen development. The function of these male-specific small RNAs remains unknown, but their different size and accumulation patterns suggest distinct biological activities. A subset of 21-nt phasiRNAs in rice is loaded onto the MEIOSIS ARRESTED AT LEPTONENE1 (MEL1) protein151, which is the ortholog of AGO5 in Arabidopsis thaliana. The mel1 mutants arrest during early meiotic stages, and produce dysfunctional pollen mother cells (PMCs) that appear frequently in developing anthers. b. ARGONAUTE (AGO) functions in meiosis, cell specification and chromosome segregation. (Left) In the female gametophyte, AGO104 in maize and AGO9 in A. thaliana were associated with non-cell-autonomous regulation of meiosis and germline specification, but the molecular pathways responsible for that are still unclear. Despite both being expressed in companion cells, AGO104 and AGO9 are involved in epigenetic silencing of transposable elements in the megaspore mother cells (MMC), perhaps through RdDM activity and mobile small RNA. (Right) Importantly, ago104 mutants also produce viable unreduced diploid gametes, indicating that AGO104 has a role meiotic chromosome segregation and establishing a direct link between small RNA regulation and apomixis. Top image in panel a adapted from 72. Right Image in panel b reproduced from 148 (arrowheads indicate micronuclei in abnormal tetrads). http://www.ncbi.nlm.nih.gov/
Meiotic Phases:
Meiosis is executed in reproductive organs in both plants and animal systems, the time and stage at which meiosis occurs varies from one system to the other.
G1 Stage: It occupies quite a large period of time at which cell prepares for the duplication of chromosomes. The necessary precursors like nucleotides, proteins and many rich molecules are mobilized. As a consequence of this accumulation, the nuclear volume increases. The chromosomes found at this stage are extremely thin and they are not clearly visible except for certain heteropycnotic chromatin regions which appear as darkly stainable segments.
S-Stage: At this stage, hitherto single stranded chromosomes undergo duplication (through DNA replication, recruitment of histone and nonhistone) into double stranded chromosomes, but the strands are held together at centromere regions. Still the chromosomes are not clearly visible. However the nuclear volume further increases.
G2 Stage: This stage is comparatively of shorter duration and cell enters into prophase. The details of molecular events are not clearly known.
Prophase I: This stage is relatively longer and most complex; basing on the behavior of chromosomes and appearance, this stage has been further divided into sub stages: sequentially these are referred to as leptotene, zygotene, pachytene, diplotene and diakinesis. In all these stages, starting with leptotene chromosomes continuously undergo condensation till metaphase.
Leptotene: At this stage, the most invisible chromosomes gradually become visible. This is due to condensation but still they appear as single stranded structure, but are actually double stranded because of chromosomal duplication takes place in the first interphase. Here, most of the chromosomes appear to be looped into horse-shoe shaped structures, where the chromosomal Telomeric ends are found to be associated with the inner face of nuclear membrane. This association has been considered as very important for some proteins that are synthesized in the cytoplasm which get associated with chromosomes through the nuclear membranes.
Chromosomes assemble axial elements (AE) along replicated sister chromatids whose ends attach to the inner nuclear membrane (NM) via a specialized conical thickening, but spermatocyte chromosomes lack the AE and the conical end thickening.
Telomeres are associated with proteins like SCP2, and the meiosis-specific cohesin STAG3 at the Sycp3-/- telomere. Chromosomal ends interact with the shelterin subunit RAP1 and the nuclear envelope protein Sun1. Attachment of telomeres to trans-nuclear envelope protein complexes facilitates to connect telomeres to motor proteins in the cytoplasm. These trans-nuclear envelope connections between telomeres and cytoplasmic motor proteins permit the active movement of telomeres and chromosomes during the first meiotic prophase. Movements of chromosomes/telomeres facilitate the meiotic recombination process, and allow high fidelity pairing of homologous chromosomes.
SUN1 and potentially SUN2, are known to be recruited to meiotic telomeres, these proteins are not meiosis-specific, therefore cannot solely account for telomere-nuclear envelope attachment and/or for other meiosis-specific characteristics of telomeres in mammals.
Research has identified a multi-subunit meiotic telomere-complex, TERB1/2-MAJIN, which takes over telomeric DNA from the shelterin complex in mouse germ cells. Shelterin subunit RAP1 is associated with the nuclear envelope protein Sun1.
Nuclear Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation shelterin subunit RAP1 with the nuclear envelope protein Sun1. http://www.ncbi.nlm.nih.gov/;http://www.ncbi.nlm.nih.gov/;http://jcb.rupress.org/
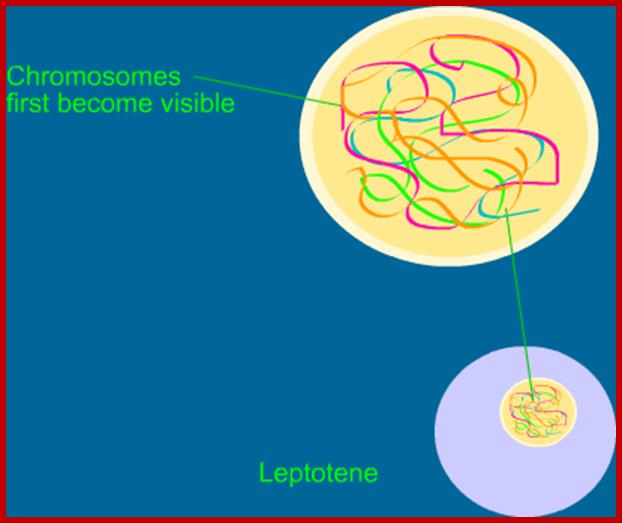
http://animalsciences.missouri.edu/biotech
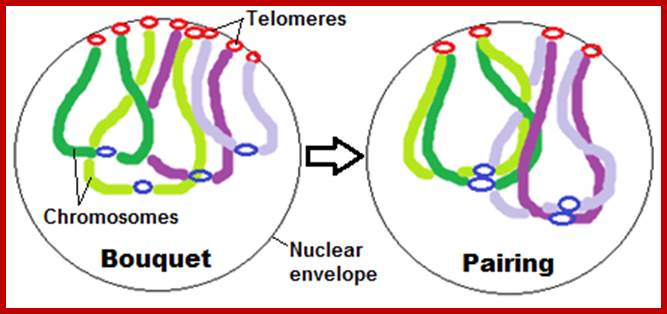
http://www.aipmtbio.co.in/
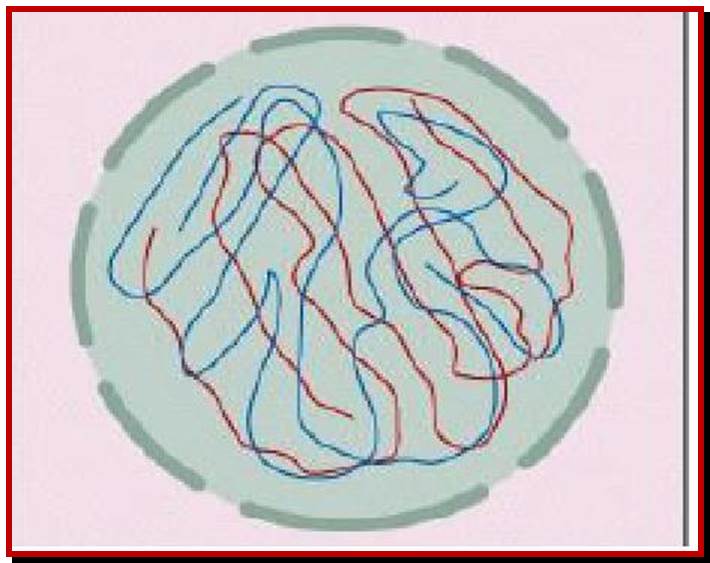
Leptotene threads already double stranded; http://classconnection.s3.amazonaws.com/
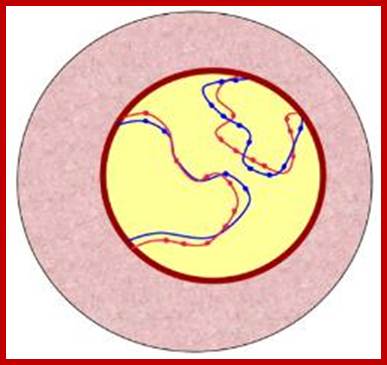
Leptotene to early Zygotene;
http://www.macroevolution.net/
This appearance of chromosomes has been referred to as “bouquet stage” where chromosomal ends ‘Telomere” are associated with inner nuclear membrane proteins; chromosomes show fine granular or bead like structures called chromomeres. They were equated to genes (by Bellings) but later they were found to be nothing but coiled expressions of chromosomes.
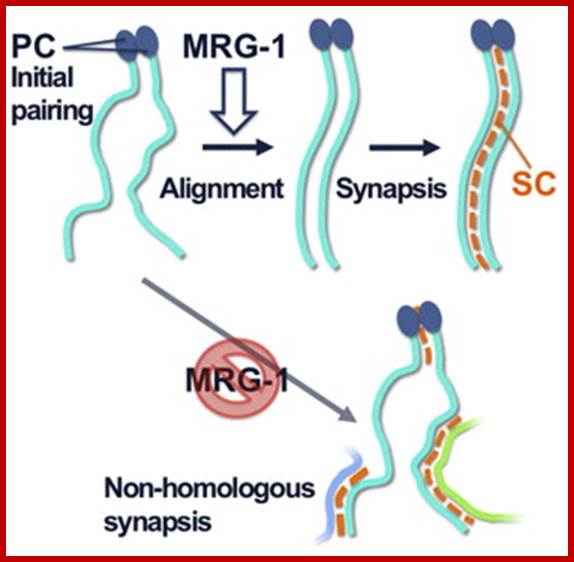
Homologous chromosome pairing is a prerequisite to establish physical linkage between homologs, which is critical for faithful chromosome segregation during meiosis I. The establishment of pairing is genetically separable from subsequent synapsis, defined as stabilization of pairing by the synaptonemal complex (SC). The underlying mechanism of presynaptic pairing is poorly understood. In the nematode Caenorhabditis elegans, a unique cis-acting element, the pairing center (PC), is essential for presynaptic pairing; however, it is not known whether and how the remainder of the chromosome contributes to presynaptic pairing. Here we report direct evidence for presynaptic pairing activity intrinsic to non-PC regions, which is facilitated by a conserved chromodomain protein, MRG-1. In mrg-1 loss-of-function mutants, pairing is compromised specifically in non-PC regions, leading to nonhomologous SC assembly. Our data support a model in which presynaptic alignment in non-PC regions collaborates with initial PC pairing to ensure correct homologous synapsis. http://www.cell.com/
-Chromo.--Pa-Ma-Pa-Pa-Ma-Ma-Ma-Pa---
-Chromo.--Ma-Pa-Ma-Pa-Ma-Ma-Pa-Ma---
Note-Chromo- chromosomes and chromosomal nucleosomes, Ma- maternal, Pa- paternal, random pairing of parental and maternal chromatids in equatorial plane; this arrangements also holds good for meiosis parental chromosomal synapsis and segregation.
Zygotene: This is a stage which homologous chromosomes (derived from paternal and maternal side), start recognizing each other and initiate pairing. The pairing may be initiated at any point, i.e. terminal, middle or anywhere but pairing is determined by homologous nucleotide sequences. However, once the pairing is initiated it proceeds like a Zipper. The process is so exact and precise; the pairing takes place– chromomere by chromomere and gene to gene. In any one of the homologous pair of chromosomes, if there is any non-homologous segments present the nonhomologous region gets looped out. If there are any segments of chromosomes which remain unreplicated, they complete replication at this stage and the DNA that is unreplicated DNA is Z-DNA for it is rich in GC content.
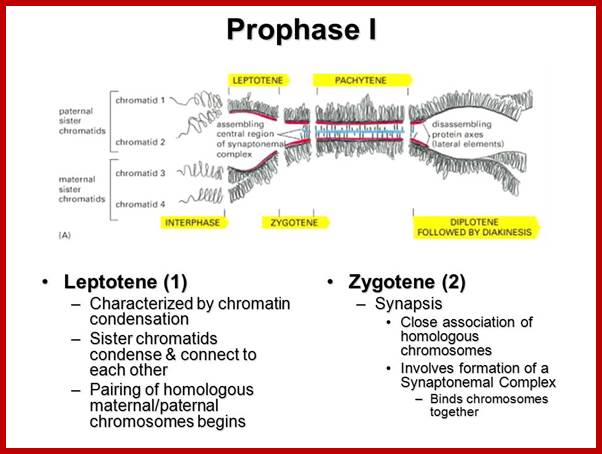
http://slideplayer.com/slide/
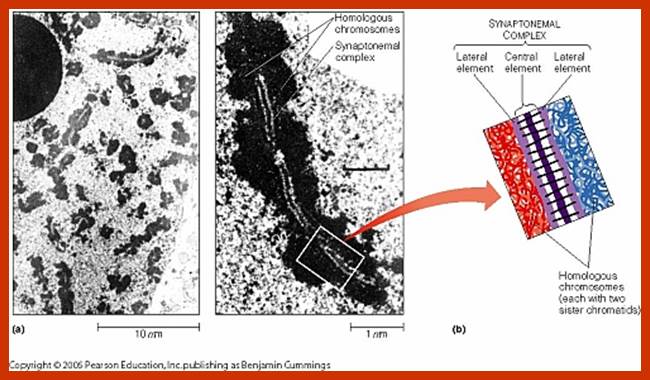
Synaptonemal elements found between meiotic prophase chromatin; www.mun.ca
Synapsis: The pairing of homologous chromosomes, though dramatic and fascinating to observe, the forces and factors that are responsible for recognizing each homologous chromosome proteins and mechanistic movement of chromosomes towards their homologous chromosomes, are still remained fully not explained. Nevertheless, electron microscopic studies of some animal cells have revealed the presence of a ribbon shaped protein complex associated with the paired homologous chromosomes. This complex has been named as Synaptonemal Complex.
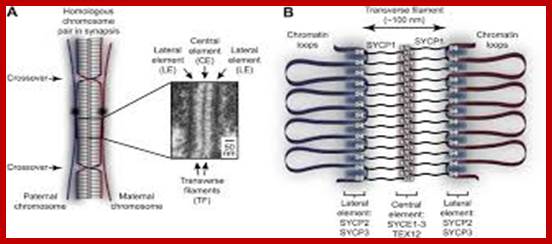
Appearance of synaptonemal elements and final synapsis;
![]()
![]()
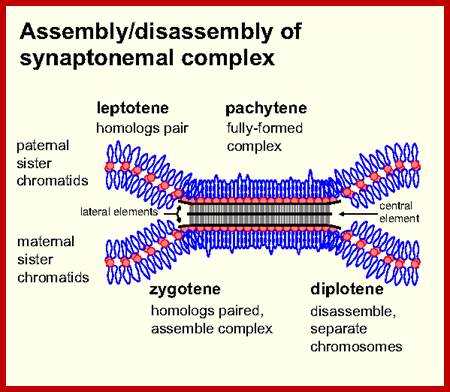
www.users.path.ox.ac.uk and http://www.carabinsnicois.fr/
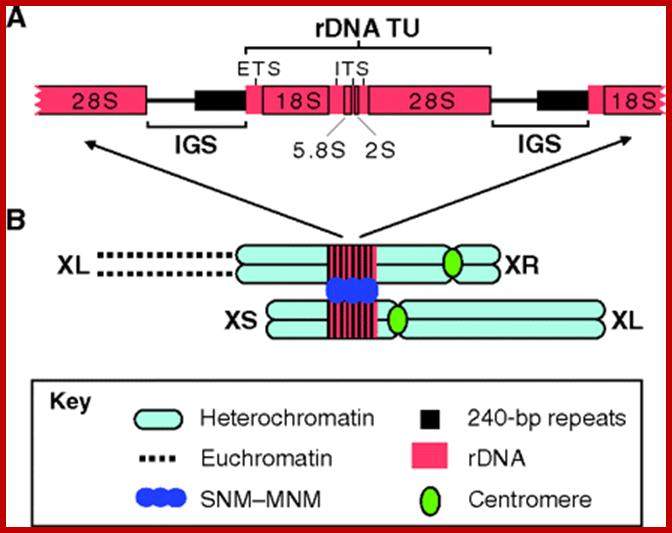
X–Y chromosome pairing in Drosophila male meiosis. (A) The rDNA transcription unit (TU) and intergenic spacer (IGS) region comprise a complete rDNA unit. Each rDNA TU consists of the 18S, 5.8S, 2S and 28S genes, the external transcribed spacer (ETS) and internal transcribed spacers (ITS). Transcription units are separated by IGSs. The IGS comprises several arrays of tandem repeats, including five to ten copies of a 240-bp repeat located immediately upstream of the rDNA TU in each rDNA repeat. (B) The X and Y chromosomes are shown schematically, with heterochromatic regions as rounded rectangles, euchromatin as dotted lines and centromeres as green ovals. rDNA loci are located in the central region of the X heterochromatin and near the base of the short arm of the Y heterochromatin. SNM and MNM are recruited to 240-bp repeats and mediate stable homologous connections, analogous to chiasmata, throughout meiosis I until anaphase I. XL, the left arm of the X chromosome; XR, the right arm of the X chromosome; YS, the short arm of the Y chromosome; YL, the long arm of the Y chromosome. Note: in diploids derived from X and Y chromosomes, as believed the X and Y chromosomes don’t pair, but they have some gene loci such as rRNA genes, which have homology, thus these loci can pair. http://jcs.biologists.org/
https://quizlet.com/
Synaptosome complex:
Structural features of synaptic elements;
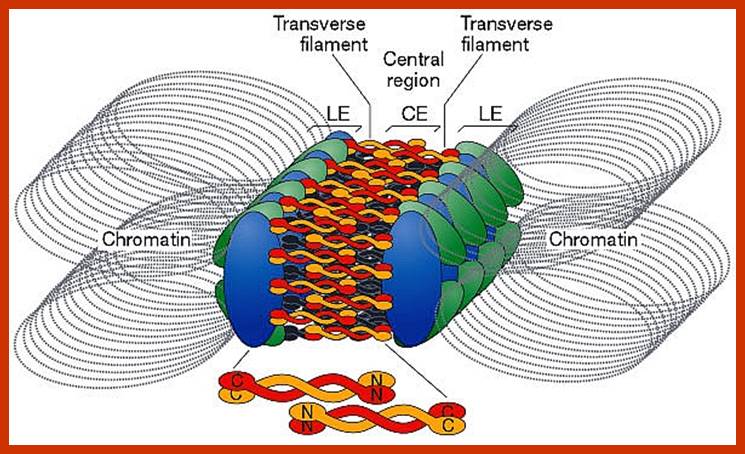
Synaptosomal elements; http://www.lookfordiagnosis.com/
This complex appears at the late leptonema or early zygonema and establishes fully at zygonema and pachynema. This structure starts disappearing at the diplonema. Synaptonemal complex is exclusively made up of protein units, arranged in between a pair of homologous chromosomes in the form of a paired ribbon. It consists of a pair of axial, a pair of lateral and a single central filament is the overlapping structure extended from the axial filaments. The space found between a pair of axial filaments; lateral filaments is 0.15µm to 2 µm. On the other side of the axial filaments, lateral filaments are present and these are associated with the homologous chromatin material. Some of the chromatin material penetrates through these protein complexes and open out into the central region as free DNA segments. These may consist of individual genes or a group of gene segments. The genetic material that is extruded from both the homologous chromosomes aligns parallel to each other, such that recombination may be brought about. Notwithstanding the above observation, the nature of the proteins and their functional mechanisms in pairing and recombination has remained as an enigma. It has been speculated that the synaptonemal complex is made up of proteins, which not only recognize the homologous segments of chromosomes, but also bring about the pairing and recombination of genetic material between them.
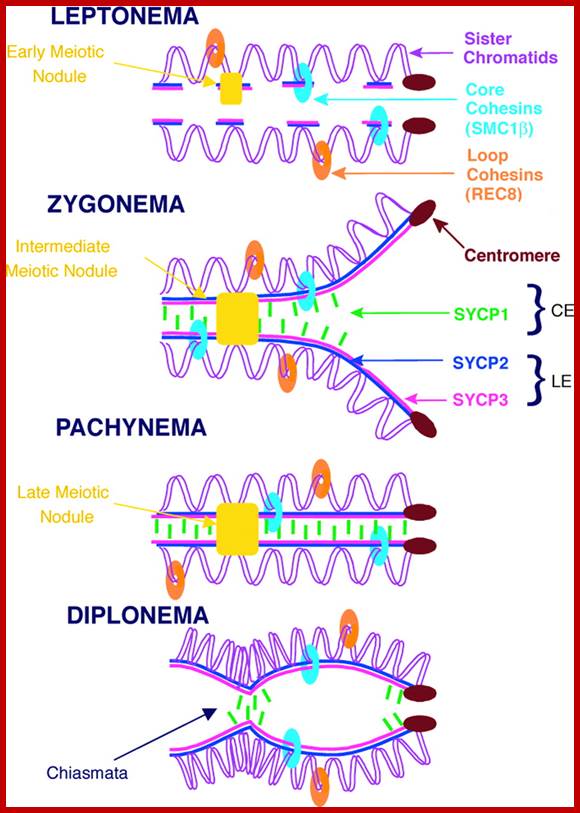
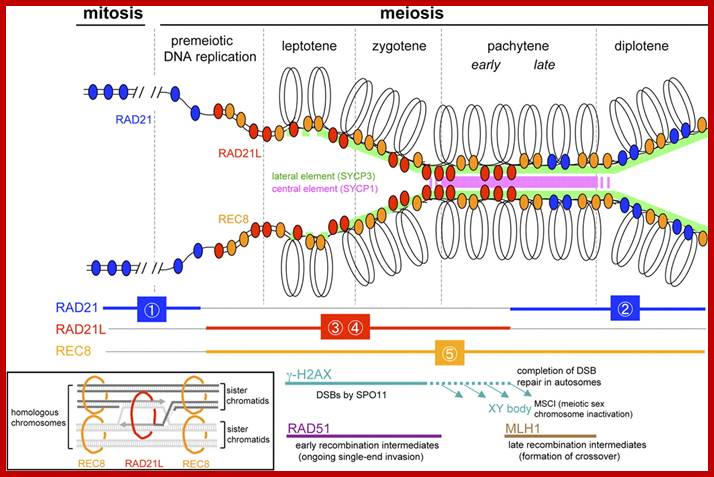
Three specific components of the synaptonemal complex have been characterized: SC protein-1 (SYCP1), SC protein-2 (SYCP2), and SC protein-3 (SYCP3). In humans, the SYCP1 gene is on chromosome 1p13; the SYCP2 gene is on chromosome 20q13.33; and the gene for SYCP3 is on chromosome 12q. www1.qoloq.com
Three specific components of the synaptonemal complex have been characterized: SC protein-1 (SYCP1), SC protein-2 (SYCP2), and SC protein-3 (SYCP3). In humans, the SYCP1 gene is on chromosome 1p13; the SYCP2 gene is on chromosome 20q13.33; and the gene for SYCP3 is on chromosome 12q. Three specific components of the synaptonemal complex have been characterized: SC protein-1 (SYCP1), SC protein-2 (SYCP2), and SC protein-3 (SYCP3). Now one more SYP4 has been added- (SYP-1, SYP-2, SYP-3, and SYP-4).
The synaptonemal complex (SC) was described by Montrose J. Moses in 1956 in primary spermatocytes of crayfish and by D. Fawcett in spermatocytes of pigeon, cat and man. As seen with the electron microscope, the synaptonemal complex is formed by two "lateral elements", mainly formed by SYCP3 and secondarily by SYCP2, a "central element" that contains at least two additional proteins and the amino terminal region of SYCP1, and a "central region" spanned between the two lateral elements, that contains the "transverse filaments" composed mainly by the protein SYCP1.
The SCs can be seen with the light microscope using silver staining or with immunofluorescence techniques that label the proteins SYCP3 or SYCP2.
This "tripartite structure" is seen during the Pachytene stage of the first meiotic prophase, both in males and in females during gametogenesis. Previous to the pachytene stage, during leptonema, the lateral elements begin to form and they initiate and complete their pairing during the zygotene stage. After pachynema ends, the SC usually becomes disassembled and can no longer be identified.
Formation of the SC usually reflects the pairing or "synapsis" of homologous chromosomes and may be used to probe the presence of pairing abnormalities in individuals carrying chromosomal abnormalities, either in number or in the chromosomal structure. The sex chromosomes in male mammals show only "partial synapsis" as they usually form only a short SC in the XY pair. The SC shows very little structural variability among eukaryotic organisms despite some significant protein differences. In many organisms the SC carries one or several "recombination nodules" associated to its central space. These nodules are thought to correspond to mature genetic recombination events or "crossovers".
In cell development the synaptonemal complex disappears during the late prophase of meiosis I.
One aspect of meiosis research (in genetics, molecular biology, and reproductive medicine) involving synaptonemal complexes is how they (or what they do) can be affected and damaged by chemical exposure, specifically to toxins like bisphenol A (an experiment at Harvard Medical School by Dr. Monica Colaiácovo, Ph.D., an associate professor of genetics, involved dosing C. elegans worms with that toxin and then verifying the results in mouse studies).
Model for organisation of the chromosome axis by SYCP3 assembly:
Each SYCP3 tetramer contains two DNA-binding regions, separated by a distance of 20 nm owing to the central rigid rod-like structure. Upon binding to the chromosome axis, SYCP3 tetramers may pinch off portions of the axis such that short stretches of chromosomal DNA are looped back on themselves with a separation of 20 nm. The loading of further SYCP3 tetramers may bridge between the initial pinched-off portions, creating a continuous structure that extends along the chromosome axis. Thus, the final assembly consists of a three-dimensional lattice of SYCP3 tetramers that organise the chromosome axis in a concertina-like manner such that the length of the axis is shortened and the chromatin loops (that will flank the SC) are lengthened. For clarity, other meiotic factors that are known to perform important functions in the organisation of the chromosome axis, such as SYCP2, Cohesin and Condensins, are not depicted; http://elifesciences.org/
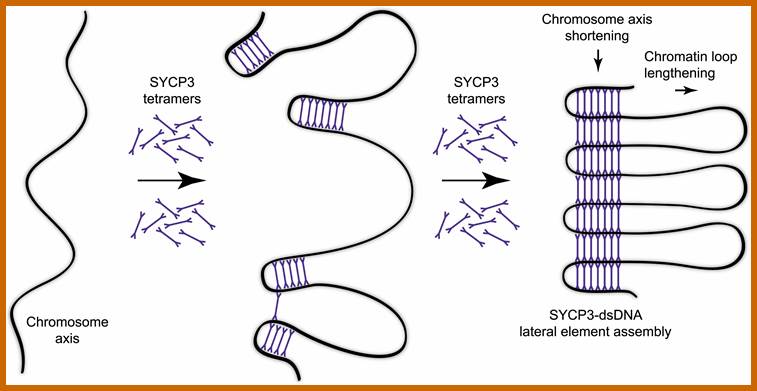
A molecular model for the role of SYCP3 in meiotic chromosome organization; http://elifesciences.org/
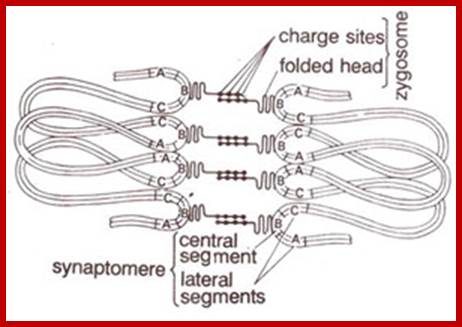
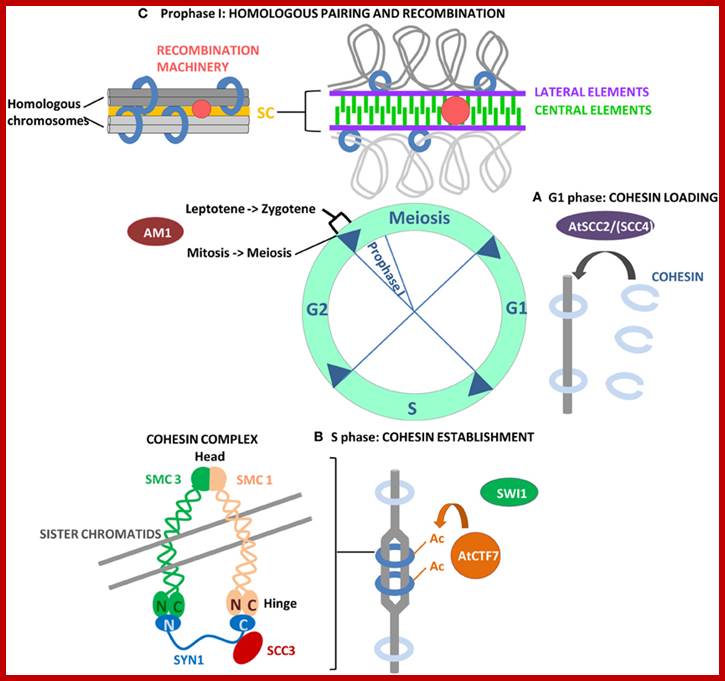
http://www.frontiersin.org/; Loading of cohesins Scca/Scc4; forming cohesion; SMC proteins are involved in this chromatin pairing.
Pachytene: At this stage, if there are any parts of homologous chromosome unpaired, the paring will be completed. Then, at certain points, the chromosomal materials between homologous pairs exchange. This phenomenon is referred to as recombination or crossing over. This involves breakage and reunion between crossing over. This involves hologous chromosomes. Use of radio-active precursors have shown that the chromosomal material, get degraded and resynthesized. The enzymes that are involved in this process have been recognized as specific endonucleases, DNA polymerases, ligases and exonucleases.
Mechanism of cross over: Basing on the microscopic observation of meiotic chromosomes, Darlington has opined that homologous chromosomes pair and coil relationally at zygotene under great strain. This coiling brings about great torsion and strain on chromonemal strands. Because of this, sister chromatids of both homologous chromatids break at these points. This breakage releases the torsion and strain; the chromatids recoil and come to rest. If the broken ends of the non sister chromatids of two homologous chromosomes are brought near to each other, then they join, thus crossing over is brought about, resulting in genetic exchange between the parental chromosomes. Thus, crossing over is often summed up as the phenomenon of breakage and reunion between two non-sister strands of two homologous chromosomes. At the gross level this appears to be true. But break down and the reunion of chromosomes involve the breakdown of chromatin materials like proteins, DNA, and the rejoining involves the resynthesis and reassembly of the chromosomal proteins.
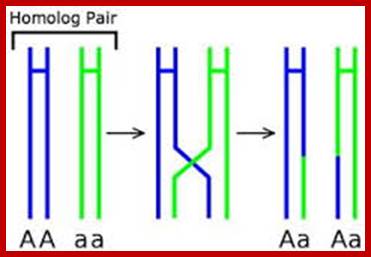
Diagram of crossing-over. Note that while crossing over is shown here, for simplicity, between only one of the two chromatids of each chromosome, each chromatid of each chromosome actually synapses with one of the chromatids of that chromosome's homolog. So crossing-over between both of the synapsed chromatid pairs does occur.www.macroevolution.net
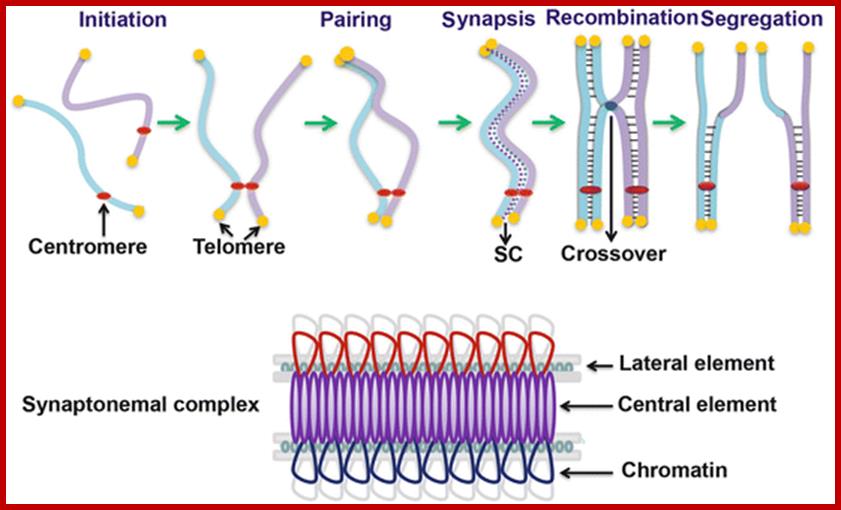
http://link.springer.com/referenceworkentry; Meiosis: Interactions Between Homologous Chromosomes; ,
Meiosis produces haploid cells to maintain the diploid genome after fusion of gametes and is essential for eukaryotic sexual reproduction. Meiosis is different to mitosis, because it involves in homologous chromosome interaction during prophase I, such as pairing, synapsis, recombination and segregation. Meiotic recombination is important for paring, synapsis and proper chromosome segregation, and also results in the redistribution of genetic materials, thereby leading to distinct genetic makeup between parents and off-springs and among individual progeny. Plants have many advantages to study meiosis genetically such as ease of access to meiotic cells and non lethality of meiotic mutations. In the past a few decades, forward genetic identification of meiotic mutants and reverse genetic studies of meiotically expressed genes in model plants of eudicot Arabidopsis and monocot rice have greatly advanced the understanding of mechanisms underlying homolog interactions during meiosis. This chapter will briefly describe the chromosomal features of meiosis and then focus on genes important for homolog interactions, as well as discussing the conservation and diversification of meiotic genes among plants and other organisms.
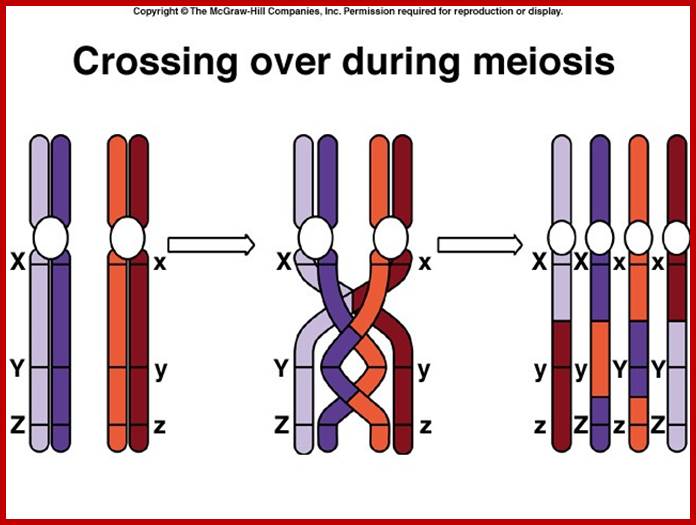
http://www.uic.edu/
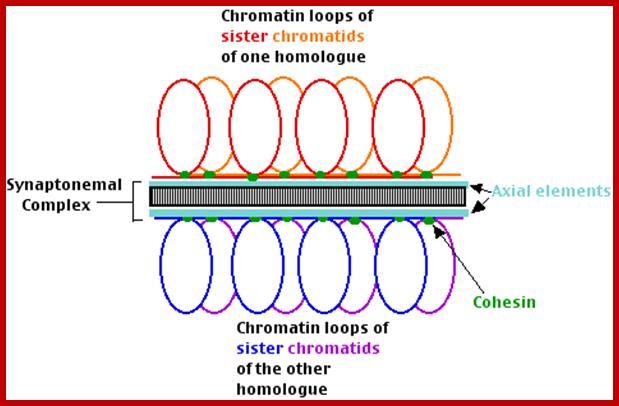
http://users.rcn.com/
http://users.rcn.com/jkimball.ma.ultranet
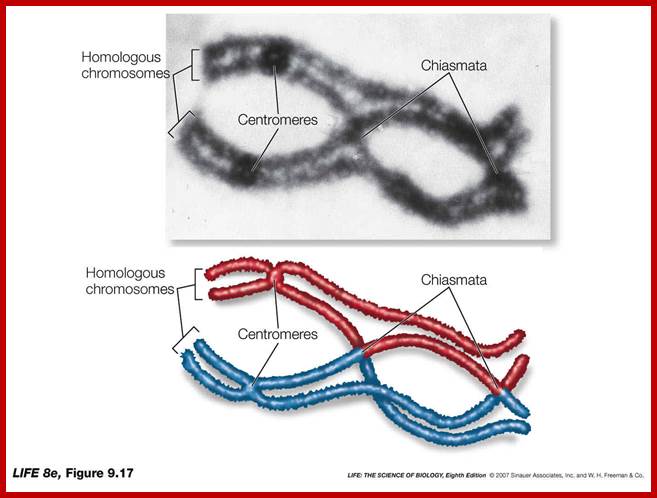
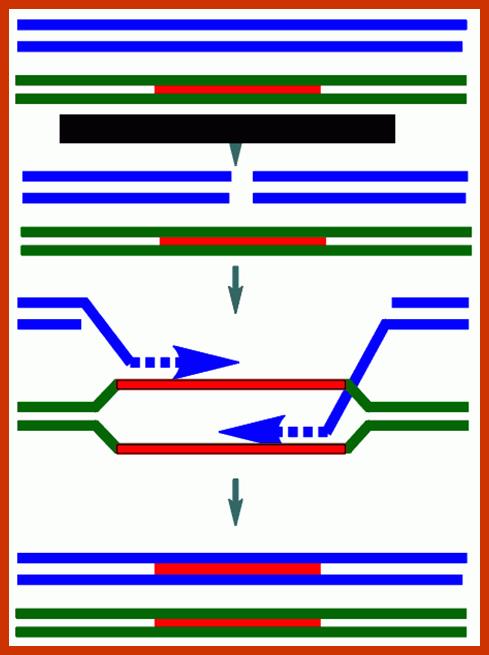
Chromosomal DNA pairing and recombination; katyperrybuzz.blogspot.com
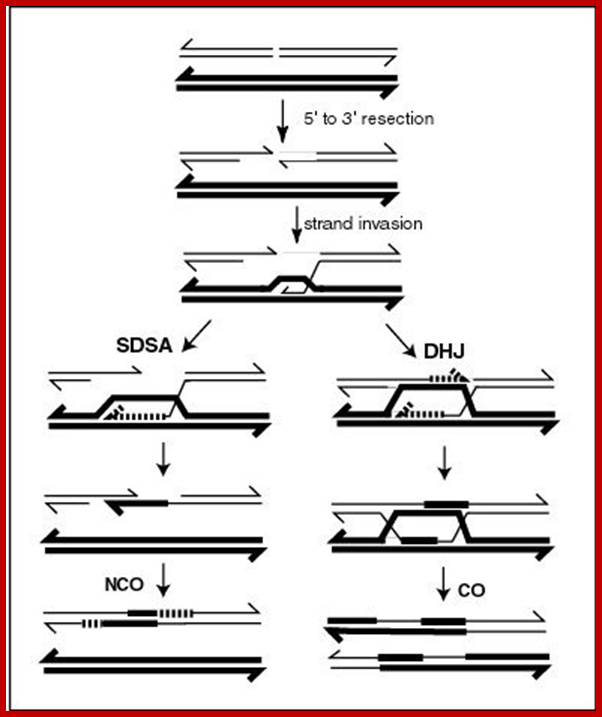
A current model of meiotic recombination, initiated by a double-strand break or gap, followed by pairing with an homologous chromosome and strand invasion to initiate the recombinational repair process. Repair of the gap can lead to crossover (CO) or non-crossover (NCO) of the flanking regions. CO recombination is thought to occur by the Double Holliday Junction (DHJ) model, illustrated on the right, above. NCO recombinants are thought to occur primarily by the Synthesis Dependent Strand Annealing (SDSA) model, illustrated on the left, above. Most recombination events appear to be the SDSA type. https://en.wikipedia.org/wiki/Genetic_recombination
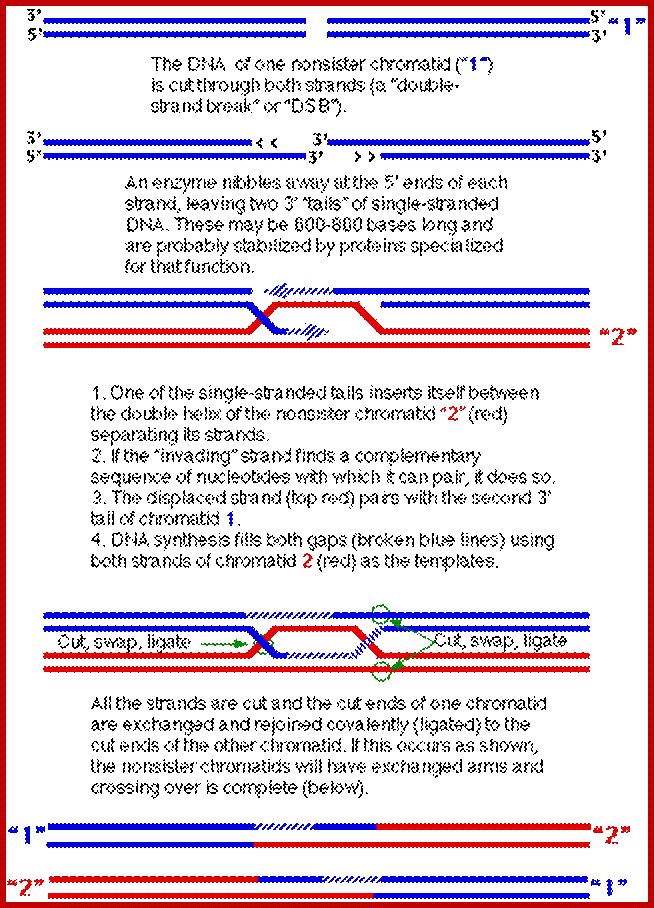
Users.rcn.com/jkimball.ma.ultranet
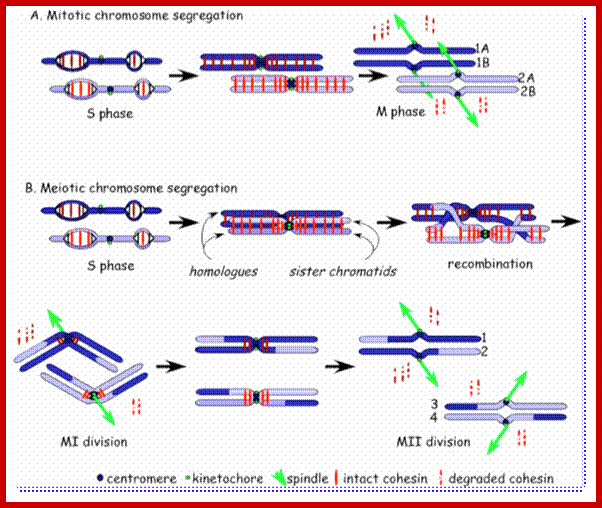
Comparison of chromosome dynamics in meiosis and mitosis: (A) Mitosis in a cell with two chromosomes ensures that each daughter cell receives a copy of each chromosome. Importantly, while the apparatus of mitosis ensures that each daughter cell will have a copy of chromosome 1 and chromosome 2, it does not distinguish which one. That is, the daughters may end up with (1A and 2A), and (1B and 2B). Or, they may end up with (1A and 2B) and (1B and 2A). Since the sister chromatids are identical, this random orientation doesn't matter. The important thing is that the daughter cells have the exact same chromosome complement as the starting cell: one copy of chromosome 1 and one copy of chromosome 2. (B) In meiosis, the starting diploid is reduced to four haploids. The homologous chromosomes are duplicated, and paired to one another. Following, recombination, the homologues separate in the MI division. The MII division separates the sister chromatids, similar to mitosis. Each daughter nucleus will receive a single chromatid from a single homologue; importantly, because of recombination, the four daughter nuclei will not be genetically identical. http://www-bcf.usc.edu/ ‘Forsburg, Mol Cell 9:703 (2002)
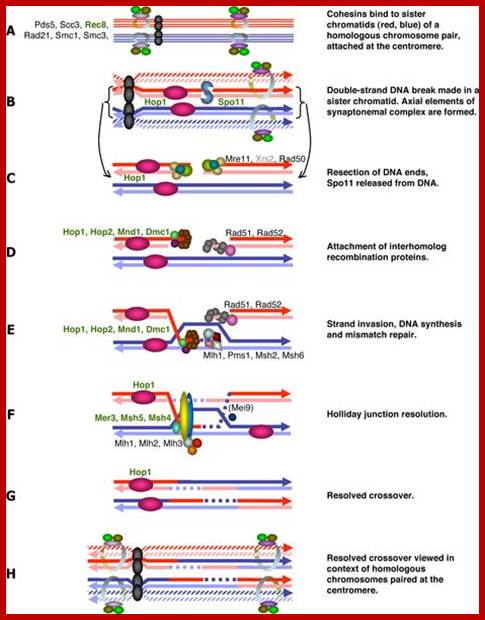
Meiotic recombination in mammals: localization and regulation http://www.nature.com/
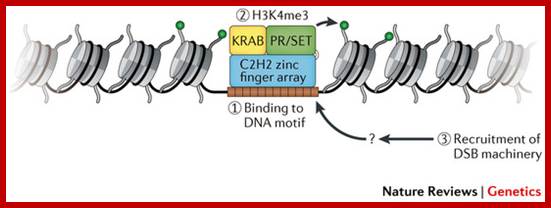
Meiotic recombination in mammals: localization and regulation http://www.nature.com/
Note that each recombinant DNA molecule includes a region where nucleotides from one of the original molecules are paired with nucleotides from the other. But no matter. The need for a smooth double helix guarantees that each exchange takes places without any gain or loss of nucleotides. So long as the total number of nucleotides in each strand and the complementarity (A-T, C-G) is preserved, this "heteroduplex"region (which may extend for hundreds of base pairs) will only rarely have genetic consequences.
And these may, in fact, be helpful because the synthesis of a short stretch of DNA using the template provided by the other chromatid also provides a mechanism for repairing any damage that might have been present on the "invading" strand of DNA. If the cut in the molecule-1 occurs in the region of a mutation, the damaged or incorrect nucleotides can be digested away. Refilling the resulting gap, using the undamaged molecule 2 as the template, repairs the damage to molecule 1.
Why should the cutting and ligation be limited to the strands shown? They are not. Half the time the cutting and ligating re-joins the original parental arms. In these cases, no crossover takes place. The only genetic change that might have occurred is a transfer of some genetic information in the heteroduplex region. So crossing over not only provides a mechanism for genetic recombination during meiosis but also provides a means of repairing damage to the genome. How and why germ cell leave mitosis and enter into meiosis. Germ cells- self renewal, another is differentiated as sperm or egg cells, leave mitotic cycle and enter into meiotic cycle.
Meiotic recombination in mammals: localization and regulation;
Frédéric Baudat, Yukiko Imai and & Bernard de Massy
During meiosis, a programmed induction of DNA double-strand breaks (DSBs) at homologous sites leads to the exchange of genetic material between homologous chromosomes. These exchanges increase genome diversity and are essential for proper chromosome segregation at the first meiotic division. Recent findings have highlighted an unexpected molecular control of the distribution of meiotic DSBs in mammals by a rapidly evolving gene, PR domain-containing 9 (PRDM9), and genome-wide analyses have facilitated the characterization of meiotic DSB sites at unprecedented resolution. In addition, the identification of new players in DSB repair processes has allowed the delineation of recombination pathways that have two major outcomes, crossovers and non-crossovers, which have distinct mechanistic roles and consequences for genome evolution.
Recombination can be homologous between sequences that are nearly equal at meiosis, or site specific- between sequences with a limited stretch of similar sequences or transposition- where DNA elements move from one site to another, where similar sequences are involved
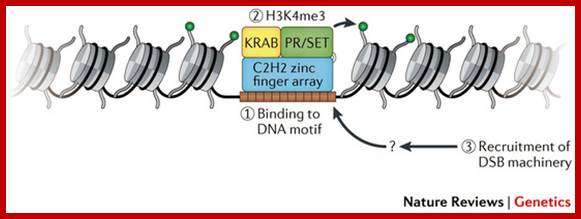
PR domain-binding 9 (PRDM9) binds to a specific DNA motif (brown squares) through its C2H2 zinc finger array (blue oblong). Subsequently, the PR/SET domain (PRDI-BF1 and RIZ homologous region, a subclass of SET domains;
Proteins involved in mammalian meiotic recombination; A | DNA double-strand break (DSB) formation requires several proteins (SPO11, MEI4, REC114, MEI1, HORMA domain-containing protein 1 (HORMAD1)) and is generated by the predicted catalytic activity of SPO11 (shown as purple spheres).
During meiosis, a programmed induction of DNA double-strand breaks (DSBs) leads to the exchange of genetic material between homologous chromosomes. These exchanges increase genome diversity and are essential for proper chromosome segregation at the first meiotic division. Recent findings have highlighted an unexpected molecular control of the distribution of meiotic DSBs in mammals by a rapidly evolving gene, PR domain-containing 9 (PRDM9), and genome-wide analyses have facilitated the characterization of meiotic DSB sites at unprecedented resolution. In addition, the identification of new players in DSB repair processes has allowed the delineation of recombination pathways that have two major outcomes, crossovers and non-crossovers, which have distinct mechanistic roles and consequences for genome evolution. http://www.nature.com/
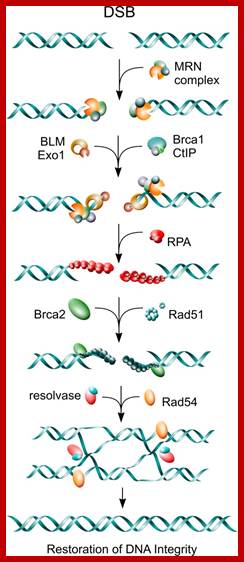
Base excision repair (BER) and nucleotide excision repair (NER) pathways. Both BER and NER repair pathways utilize the complementary DNA strand to restore sequence information lost in the damaged DNA strand. A) Schematic representation of the basic steps followed during short-patch BER (see text for details). B) Main sequence of events and enzymatic activities implicated in NER. C) Forms of lesions generated in the DNA by IR. Emil Mladenov and George Iliakis; http://www.intechopen.com/
Molecular events during recombination; www.useek.com
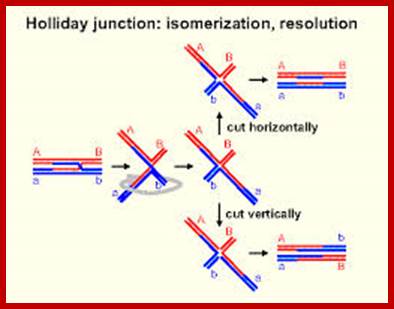
Molecular Mechanism of crossing over: When homologous chromosomes are brought together or paired by synaptonemal complex, some of the segments or chromosomal regions of homologous chromosomes get disassociated with chromosomal proteins and the DNA strands from the apposing chromosomes loop out with the central region found between two axial filaments of SC.
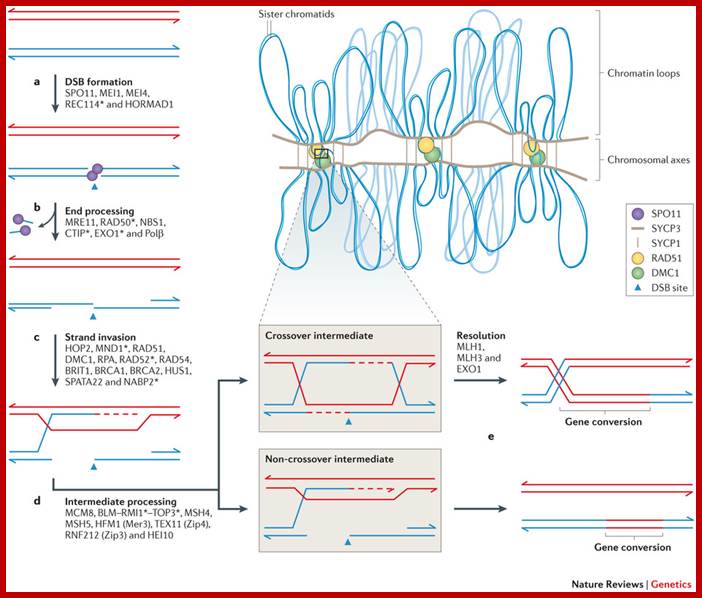
During meiosis, a programmed induction of DNA double-strand breaks (DSBs) leads to the exchange of genetic material between homologous chromosomes. These exchanges increase genome diversity and are essential for proper chromosome segregation at the first meiotic division. Recent findings have highlighted an unexpected molecular control of the distribution of meiotic DSBs in mammals by a rapidly evolving gene, PR domain-containing 9 (PRDM9), and genome-wide analyses have facilitated the characterization of meiotic DSB sites at unprecedented resolution. In addition, the identification of new players in DSB repair processes has allowed the delineation of recombination pathways that have two major outcomes, crossovers and non-crossovers, which have distinct mechanistic roles and consequences for genome evolution. Frédéric Baudat et al; http://www.nature.com/
Genetics of Meiosis:
Meiosis leads parental chromosomal segregation randomly thus gametes produced contain parental chromosomes not evenly segregated. Thus, one can see meiosis leads segregation of two independent genes as we find in the diagram below.
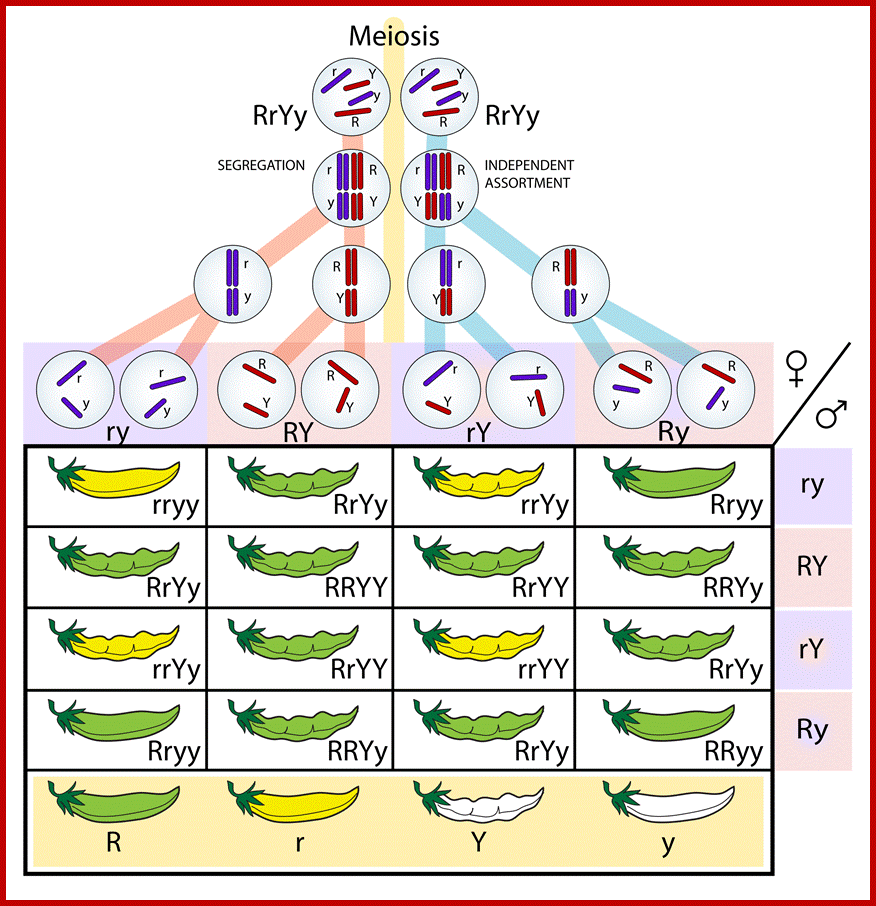
En.wikipedia.org
Germ cells face a number of major fate decisions during their development. One is self-renewal or differentiation; another is how to differentiate as a gamete of a particular sex, in animal sperm or egg; and a third is a cell cycle decision—to leave the mitotic cell cycle and enter the meiotic cell cycle, or more simply, the mitosis/meiosis decision.
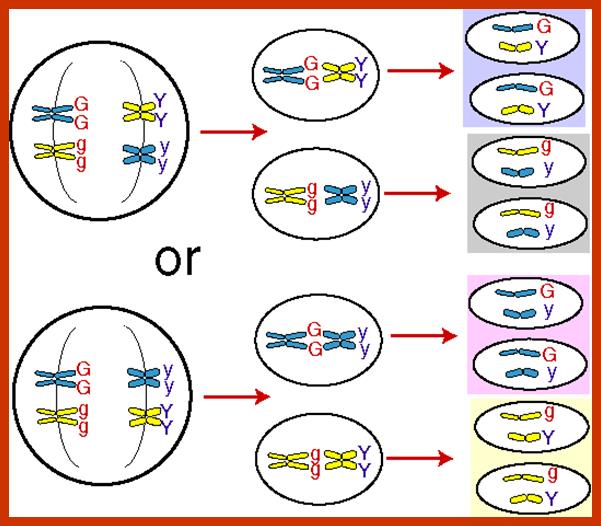
Chromosomal segregation and events at which genes segregate or assort
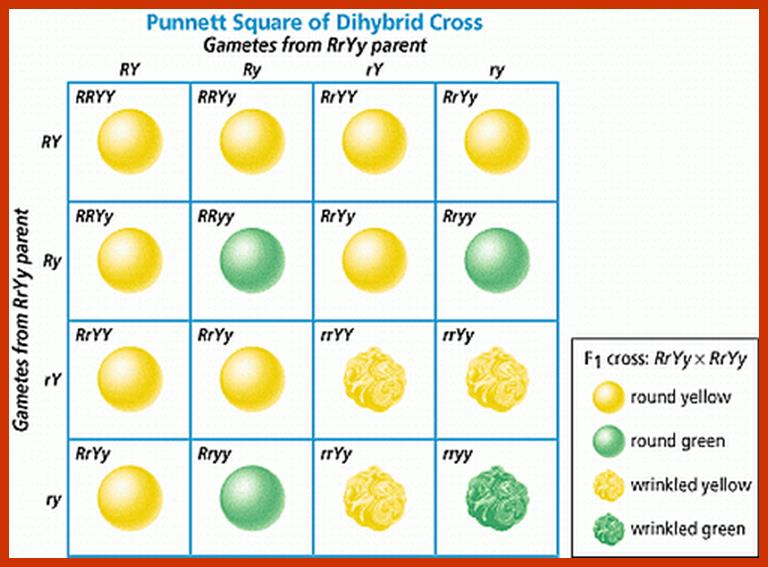
Factors segregate and assort
Neurospora spore formation and genes assortment
Importance of Meiosis;
- It acts as an antithesis for fertilization. Genetic recombination takes place between parental homologous chromosomes.
- As parental homologous chromosomes arrange randomly at first metaphasic plate, chromosomes segregate randomly but independently.
- Genetic recombination and random segregation result in the production of gametes with variable genotype.
- Variable genotypes help in bringing about variation result in the production of gametes with variable genotype.
- This variation plays a significant role in evolution.
Mitosis |
Meiosis |
|
1. It is an equational cell division |
It is a reductional cell division. |
|
|
|
|
2. Chromosome number is maintained between cell generation. |
Chromosome number is reduced to half the original number |
|
|
|
|
3. If helps in the growth of the population of cells and it takes place during growth and development |
It takes place during gameto genesis. |
|
|
|
|
4. This process takes place at 5 different stages viz. Interphase, prophase, Metaphase, Anaphase and Telophase. |
This process takes place at two stages called Meiosis I and Meiosis II. Prophase I is further sub-divided into Leptotene, Zygotene, Pachytene, Diplotene and Diakinesis. |
|
5. Homologous chromosomes do not pair, do not exchange chromosomal segments by crossing over. |
Homologous chromosomes pair and undergo crossing over. |
|
|
|
|
6. Homologous chromosomes do not undergo segregation. |
They undergo random but independent segregation. |
|
|
|
|
7. The daughter cells produced are qualitatively and quantitatively similar and do not exhibit any variation |
The daughter cells produced are qualitatively ad quantitatively dissimilar and exhibit variation. |
Meiosis is derived from Mitosis, when plants and animals started producing gametes and spore. In sexual reproduction diploids have to produce haploids as gametes or haploid spores. During meiosis many errors cause genetic variations among the off springs. Errors can happen during recombination, or during segregation. Visualize, in women the gamete formation calls remain for more than 45. During such periods any variation is possible
Meiosis and the Cell Cycle;
The special behavior of the chromosomes in meiosis I requires some special controls. Nonetheless, passage through the cell cycle in meiosis I (as well as meiosis II, which is essentially a mitotic division) uses many of the same players, e.g., MPF and APC. (In fact, MPF is also called maturation-promoting factor for its role in meiosis I and II of developing oocytes.