PLANT ENERGY TRANSFORMATIONS 2
Photosynthesis:
When life originated on this planet some 3.8 billion years ago, the first life forms were single celled heterotrophs. They were depending upon both the organic and inorganic compounds available in ‘Mother Ocean’ fresh and sweet water. With time, ‘organic soup’ created in earlier years, almost exhausted by the fast growing and multiplying population of then existing organisms. There was an imminent danger for the survival of heterotrophs. At this juncture, new life forms arose and they had the unique potentiality to capture solar energy and use the same for the synthesis of simpler organic compounds where the solar energy is stored in the form of chemical bond energy. The origin of photosynthetic organisms saved the heterotrophic organisms by providing food materials to them. Photosynthesis also provided oxygen and food; the present oxygen level is due to photosynthesis. At the same time, they also provided an environment for the diversification and sustenance of organisms. It is these organisms that produced oil fossil “fuel reserves” found all over the world; thankless are those few on this planet who are exploiting the reserves and getting richer and richer.
www.plantcell.org.uk
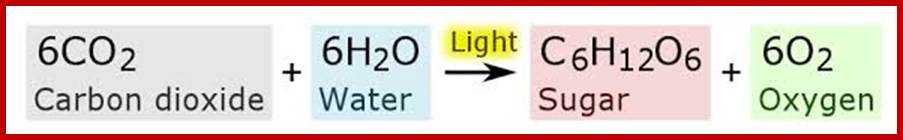
In the light of knowledge gained in recent years, the process of photosynthesis is defined as “a series of processes in which green plants capture, convert and conserve solar electromagnetic energy in the form of chemical energy call them as chemical bonds in organic compounds. Compare this to solar panels used for solar energy converting into electrical energy but plants convert solar energy into chemical energy and stored I carbohydrates; the food of all animals on this planet.
Light from milky way; http://www.bio.miami.edu/
http://www.bio.miami.edu/
http://eesc.columbia.edu/
Chemical Equation of Carbon Fixation; http://commons.wikimedia.org/
6CO₂ +12H₂O-->C₆H₁₂O₆ + 6H₂O +6O₂
6∆H(CO₂) +6∆H (H₂O)-∆H(GLU)- 6∆(HO₂) +E (SR) – E (IR) = 0
Overview of Photosynthesis and Respiration; Free energy Cycle: www.themeister.co.uk
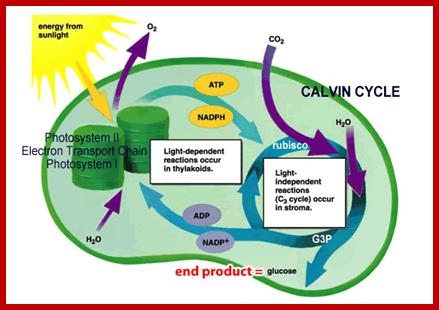
Overall process of photosynthesis in chloroplasts; reactions in grana and stroma; www.biologycorner.com
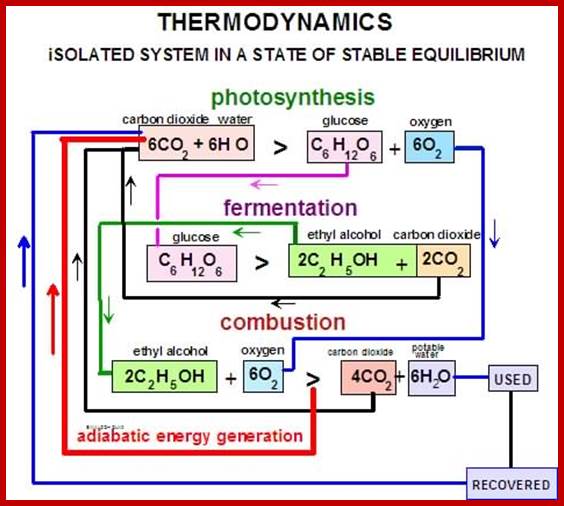
Thermodynamics of Photosynthesis; www.miller-carbon-cycle.org
Our; (Erik Albarrán-Zavala * and Fernando Angulo-Brown ) thermodynamic analysis of photosynthesis starts by establishing the following convenient working hypothesis: a) The Sun, the Earth and the Photosynthetic organism (PO) are three different thermodynamic systems. b) The Sun is a thermal reservoir with constant temperature TS = 5762 K. c). c) The Sun has constant pressure, volume and chemical composition. d) Earth behaves as a thermal reservoir at TE = 298.15 K. e) The Earth is a system with constant pressure, volume and chemical composition. f) The photosynthetic organism (PO) has constant pressure, volume and temperature, with TPO = TE = 298.15 K. g The PO chemical composition is not constant. h) All photosynthetic reactions are isothermal processes at TPO = 298.15 K.
Sun ----à PO ---àEarth
Ѵ
Chemical work (overall energy fluxes)
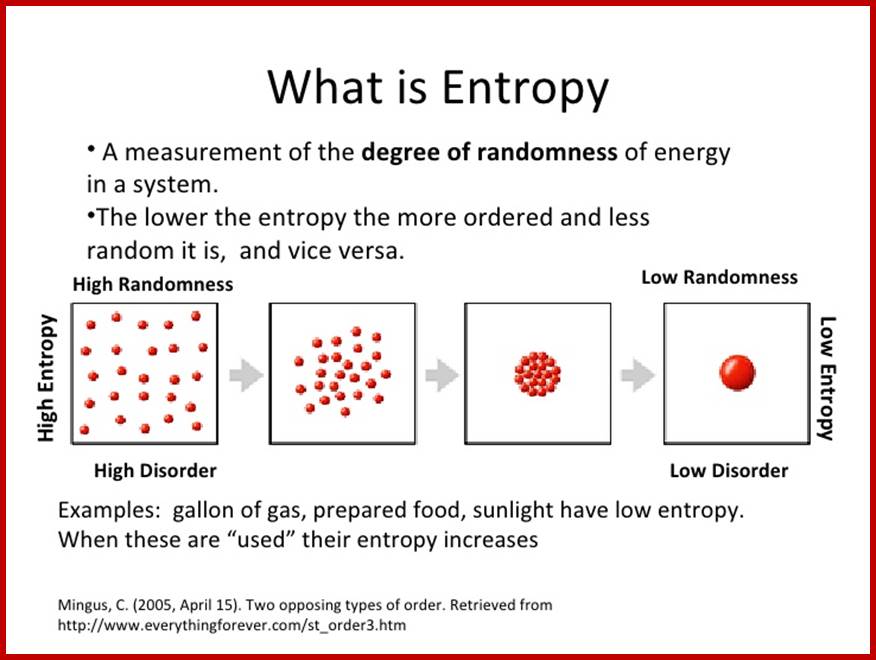
Energy, Entropy the second law of thermodynamics and how it relates to the Environment; simple graphic expression; http://image.slidesharecdn.com/
Zeroth law of thermodynamics: If two systems are both in thermal equilibrium with a third system then they are in thermal equilibrium with each other.
First law of thermodynamics: The law of conservation of energy states that the total energy of an isolated system is constant; energy can be transformed from one form to another, but cannot be created or destroyed.
∆U=Q-W
Change in the internal energy = Heat added to the system – work done by the system
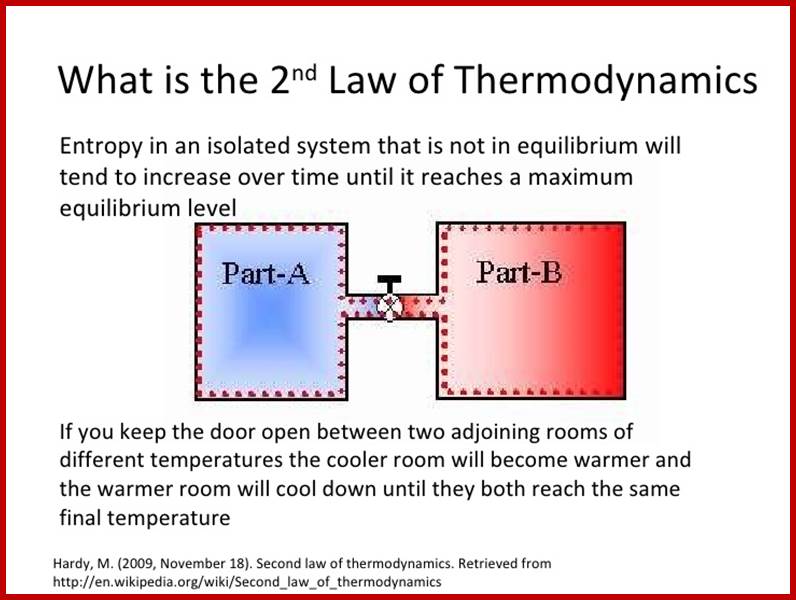
2nd law of Thermodynamics- Entropy in an isolated system that is not in equilibrium will tend to increase over time until it reaches a maximum equilibrium level;
3rd law of
Thermodynamics: The entropy of a
perfect crystal of any
pure substance approaches zero as the temperature approaches absolute zero.
http://image.slidesharecdn.com/
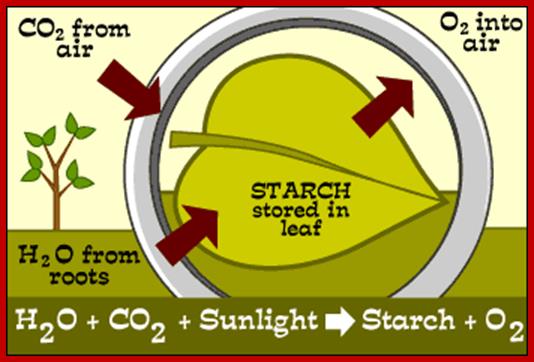
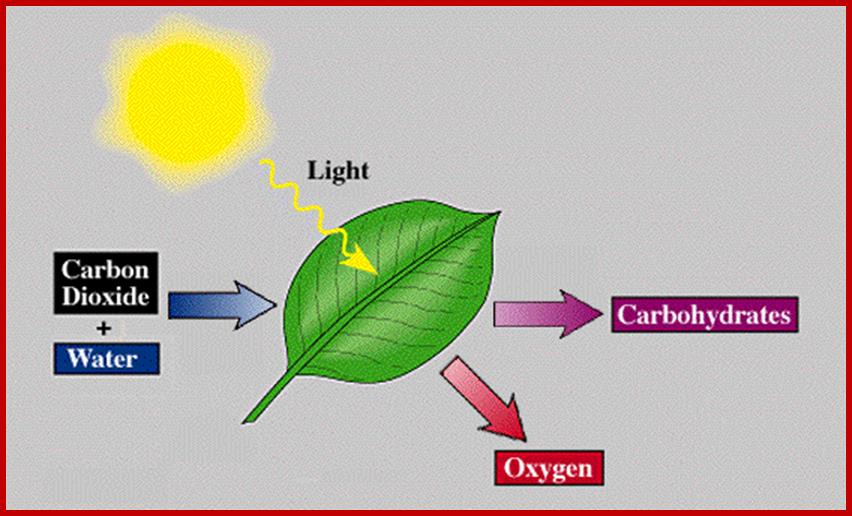
Inputs and Outputs of photosynthesis; www.phschool.com
Sun Light emits rays which contain photons, each photon carries energy depends upon the wave length-Red photons of light carry about 1.8 electron volts (eV) of energy, while each blue photon transmits about 3.1 eV. Energy of one photon of blue light that has a wavelength of 425 nm and red light that has a wavelength of 740 nm. Determine which photon has the highest energy.
https://homework.study.com/
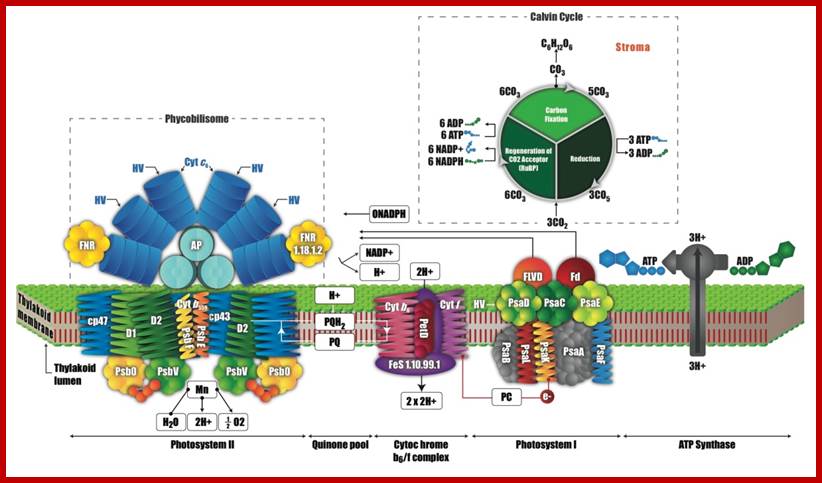
Analyzing Data for Systems Biology: Working at the Intersection of Thermodynamics and Data Analytics; Cannon, W.R., and Baxter, D. J; https://wrcannon.com
HISTORICAL REVIEW:
Ever since man learnt the art of agriculture, he understood the importance of light in the development of green plants and crops. Nevertheless, it was left to the great Aristotle, a philosopher who noted the relationship between light and greening of plants. Later, Joseph Priestley demonstrated that green plants are capable of purifying air by liberating oxygen, which is called dephlogisticating. Considering the purifying effects of plants on atmosphere, Jan Ingenhauze, working alone for a decade or so, came out with a conclusion that light i.e., solar energy is absolutely essential for the green plants to purify the air. It almost amounted that green plants liberate oxygen in the presence of light.
· Green Plants ->O2 (J. Priestley)
· Green Plants + light ->O2 (J. Ingenhauze)
· Later Jean Seedier (1782), a Swiss Pastor, pointed out that the fixed air (i.e. CO2) is very essential for the purification of air, for oxygen that is liberated during photosynthesis comes from CO2.
· Co2 + light -> C+O2 (Jean Senebier)
· Nicholas Theodore de Saussure (1804) elucidated the process of photosynthesis in which carbon combines with H2O to produce organic matter. The gain in organic matter, during the growth of green plants in sunlight is due to this process.
· C+H2O Green plants (CH2O) n (Nicholas De Saussure)
· CO2 + H2O light energy (CH2O) n (Robert Mayer)
· Dutrochet (1837) clearly demonstrated the relationship between photosynthesis and green colored chlorophyll. However, Julius Robert Mayer (1845) came out with a view that during photosynthesis, plants capture fleeting solar energy particles and store it in the form of chemical energy in organic matter. Sachs (1864), who was a pioneer in experimental botany, demonstrated that the ultimate product of photosynthesis starch.
Co2+H2O+light energy/green plants = starch
· Few decades later, Robert Hill (1937) demonstrated that the oxygen liberated during photosynthesis comes from H2O and not from CO2. For the first time radioactive isotopes were used to demonstrate the source of O2 during photosynthesis in isolated chloroplasts. The splitting of H2O into hydrogen and oxygen was then called as Photolysis. Later studies revealed that the splitting of water is not due to photolysis but it is due to photo ionization. This process is called Hill’s Reaction.
H2O+ light / Chloroplast -> (H) + OH
4OH - > 2H2O + O2
· Von Neil showed similar reactions in photosynthetic bacteria, where the hydrogen source supplied was not H2O but H2S.
· H2S + light = (CH2O) +S (von Neil)
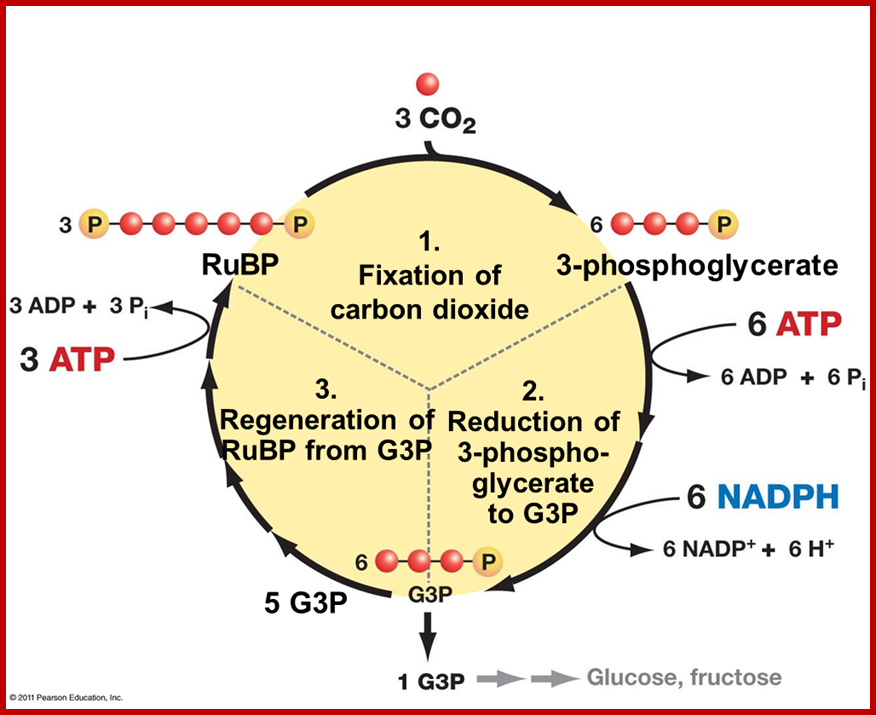
· Though Lei big (1845) and Blackman (1905) showed the importance of CO2 and other factors essential for photosynthesis, it is Melvin Calvin and his associate Benson (1940-45) elucidated the pathway of carbon fixation. This was a remarkable piece of work for which Calvin was awarded a Nobel Prize. Since then, a large number of workers have contributed their might in understanding and elucidating many facts of this complicated process.
PHOTOSYNTHETIC ORGANELLE
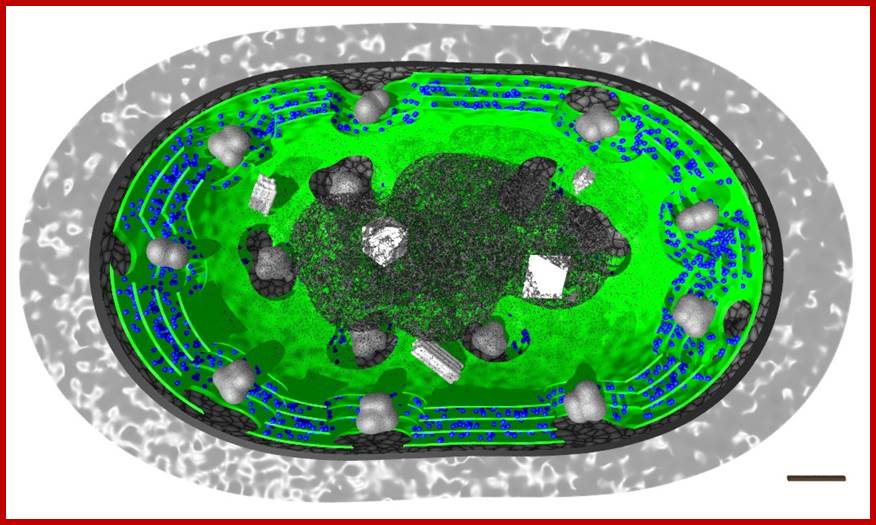
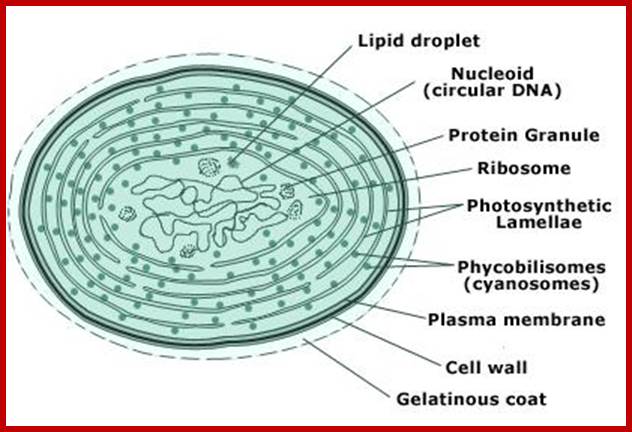
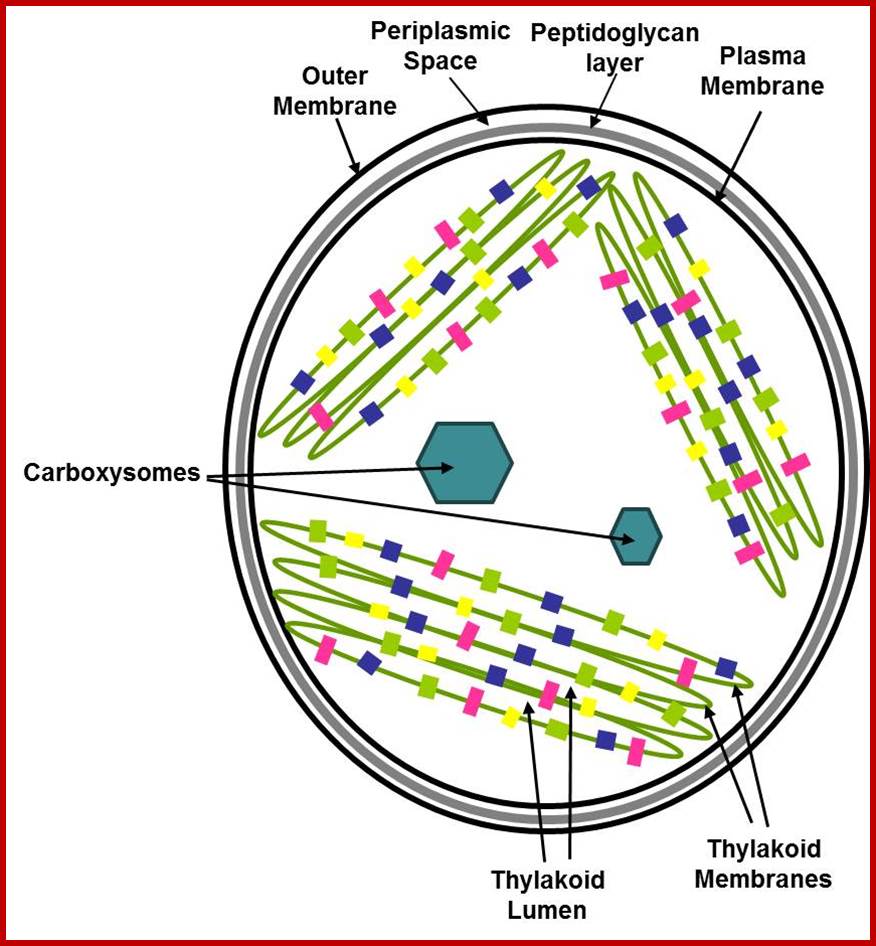
With the exception of fungi almost all-eukaryotic plants contain well-developed chloroplasts. The color of plastids depends upon the dominance of specific type of pigments. Prokaryotic blue green algal cells contain thylakoid-granal membranes embellished with photosynthetic membranes with required Quantasomes like structures for the absorption of light and performing light reactions.
Eukaryotic chloroplasts are derived from unicellular blue green algal cells by engulfing the tiny Green cells and establishing symbiosis. They have lost independent living but retained some and dependent on host cells. Eukaryotic plastids are bound by two-unit membranes within which liquid is found, it is called stoma or stromatic fluid. Suspended within the stroma there are a number of highly organized membranous structures called Grana. Cyanophycean cells are almost like Plastids.
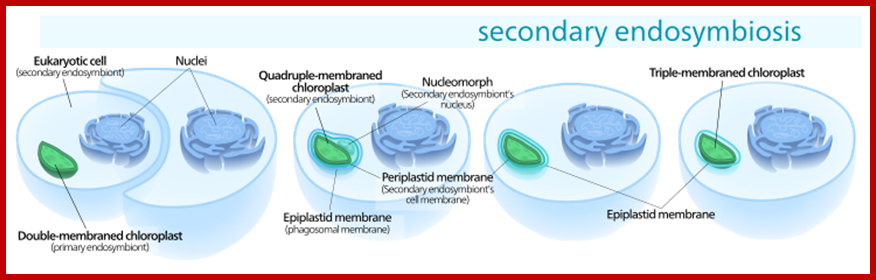
Secondary endosymbiosis consisted of a eukaryotic alga being engulfed by another eukaryote, forming a chloroplast with three or four membranes; https://en.wikipedia.org
Cyanophycean Algae:
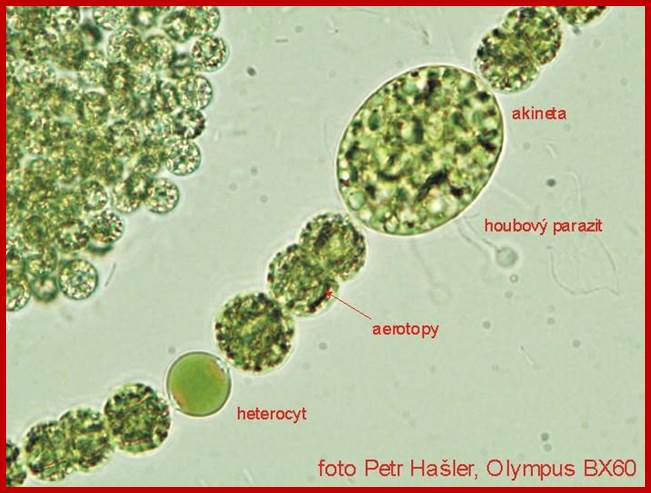
 The Thylakoid Membranes of
Nostoc cyanobacteria; photosynthetic protein complex known as photosystem II
resides in the thylakoid membranes (shown here in bright green) of plants,
algae and cyanobacteria. Suga et
al.1 report a
high-resolution X-ray crystal structure of photosystem II from the
cyanobacterium Thermosynechococcus
vulcanus.
The Thylakoid Membranes of
Nostoc cyanobacteria; photosynthetic protein complex known as photosystem II
resides in the thylakoid membranes (shown here in bright green) of plants,
algae and cyanobacteria. Suga et
al.1 report a
high-resolution X-ray crystal structure of photosystem II from the
cyanobacterium Thermosynechococcus
vulcanus.
In the above pictures only the photosynthetically active components have been marked in color: thylakoid membranes in green and phycobiliproteins in blue color. http://www.scivit.de/
Cyanophycean cell; www.gopixpic.com
https://newunderthesunblog.wordpress.com
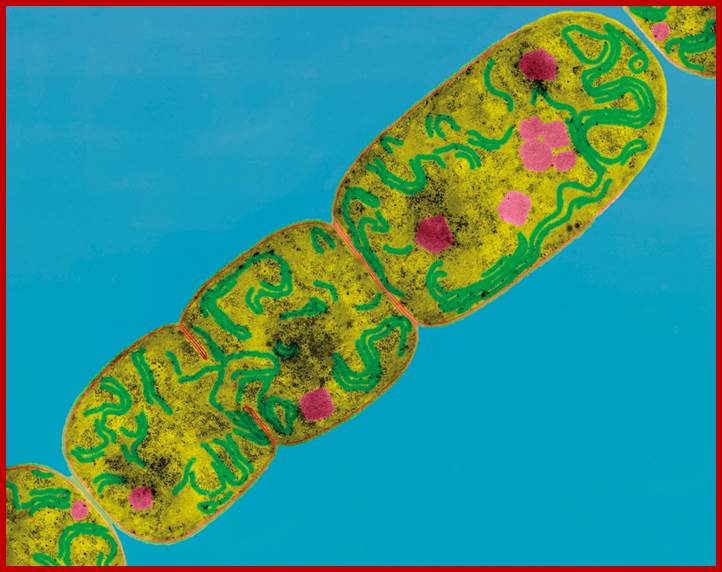
Bacterial Chlorosome:

Electron microscopic tomogram of dividing cell of the green bacteria, Chlorobaculum tepedium, with chlorosomes rendered in simulated colors; http://science.energy.gov
Bacterial Chlorosome is a solar energy harvesting system. It is the most stable structure; it is also one of the most efficient light-harvesting systems in nature. Chlorosomes were discovered in 1964 in Chlorobi species.
Bacteriochlorophyll and carotenoids are two molecules responsible for harvesting light energy. Current models of the organization of bacteriochlorophyll and carotenoids (the main constituents) inside, the Chlorosomes have lamellar organization, where the long farnesol tails of the bacteriochlorophyll intermix with carotenoids and each other, forming a structure resembling a lipid layers stacked one above the other. Chlorosomes are found to be very efficient in light harvesting. Silicon solar cells have common structure and functions.
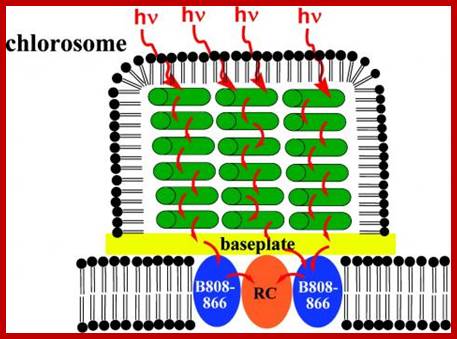
Light
harvesting antenna in bacterial ’Chlorosome’-Bacterial solar energy harvesting
‘Chlorosome” (shown in green) capture and transfer light energy to the reaction
center for photosynthesis in bacteria. New research from Oak Ridge National
Laboratory reveals that the Chlorosome maintain their structure even under
extreme conditions. http://phys.org/
Structural studies of some of nature's most efficient light-harvesting systems are lighting the way for new generations of biologically inspired solar cell devices. "It's one of the most efficient light harvesting antenna complexes found in nature," said co-author and research scientist Volker Urban of ORNL's Center for Structural Molecular Biology, http://phys.org/
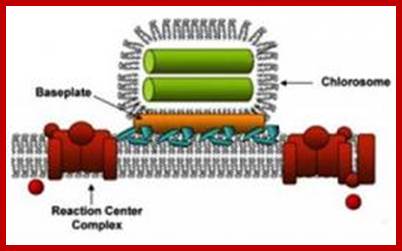
The
photosystem in green bacteria consists of a light-harvesting antenna called a Chlorosome
and a reaction center. The energy of the light the pigments absorb is
transferred to the reaction center (red) through a protein-pigment antenna
complex called the baseplate (gold). The antenna (green) is made of rod-shaped
aggregates of pigment molecules. Credit: Blankenship/WUSTL;
Read more at: http://phys.org/news/2011-11-light-harvesting-antenna.html#jCp;
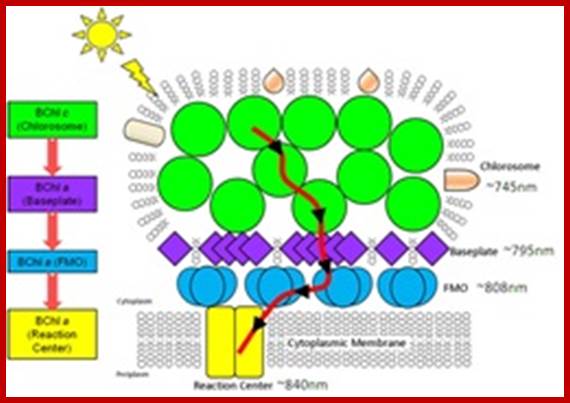
Structural model of green bacterial chlorosome showing pathway of energy transfer; http://science.energy.gov/
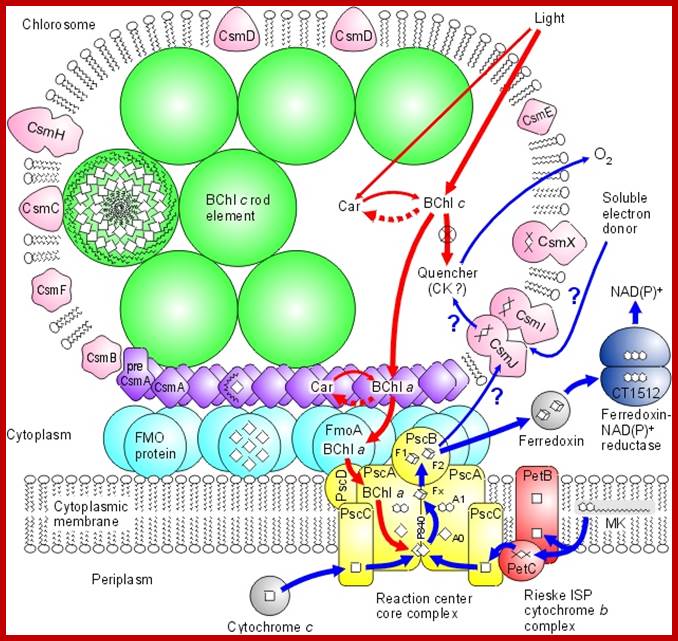
Model of the photosynthetic apparatus in green sulfur bacteria; Chlorobium tepedium shows a model of how the Chlorosome and other components of the photosynthetic apparatus are arranged in and around the cytoplasmic membrane of the green sulfur bacterium Chl. tepedium. The Chlorosome (Fig.) is unusual because of its large size and because its main antenna pigment, BChl c, is not organized on a protein scaffold but is "self-organizing" by molecular aggregation. http://www.bio.ku.dk/
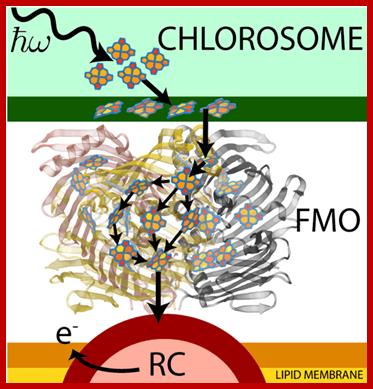
http://www.ks.uiuc.edu/Research/fmo/
Green sulphur bacteria harvest sunlight by absorbing solar energy in large pigment-containing vesicles known as a chlorosomes and transporting this energy to a reaction center for charge separation. Along the way, excitation absorbed by the Chlorosome is passed through the Fenna-Matthews-Olson (FMO) complex to get to the reaction center complex. FMO-homo-trimer from Chlorobaculum tepidum; Each monomer sandwiches 7 bacteriochlorophyll molecules for light harvesting. Between each monomer there is an additional 8th bacteriochlorophyll to make a total of 24 for the trimer. The conventional numbering scheme is shown. As this process occurs in a biological environment, there is a significant amount of thermal noise present. As excitation is passed between pigments, from the bacteriochlorophylls (BChls) in the chromosome to those in FMO and finally to those in the reaction center (RC), it is constantly under the influence of thermal fluctuations. It is of importance then that FMO can efficiently conduct excitation energy, i.e., without much energy loss. One way to do this could be by exploiting quantum coherence to speed up energy transfer. http://www.ks.uiuc.edu/
Each chlorosome consists of 250,000 pigments that capture light energy and transfer to the reaction center (base plate) where ATPs are generated. Light harvesting complexes or Antennae consists of many pigment molecules associated with protein complexes to absorb light energy of photons at a particular wave length and pass the energy on to reaction centers. In reaction centers collected energy is used in synthesizing ATP.
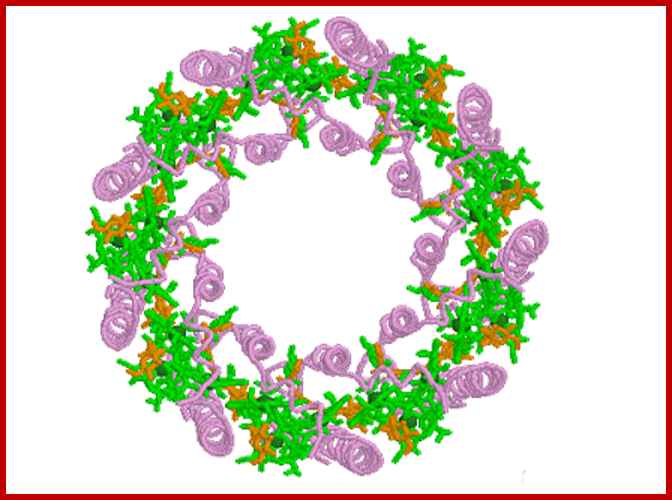
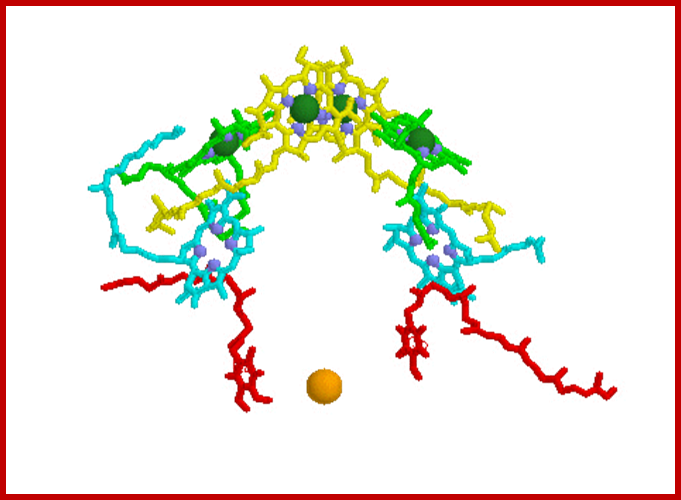
Bacterial reaction centers, a dimer yellow with green Mg2+, surrounded by several species of protein-pigment complexes know n as light harvesting chlorophylls (above diagram); Bacterial reaction /light harvesting complex; https://www.biologie.uni-hamburg.de/ http://www.chm.bris.ac.uk/
Phycobilisome at light harvesting complex in Chlorosome; http://www.chm.bris.ac.uk/
EUKARYOTIC PHOTOSYNTHETIC ORGANELLE;
With the exception of fungi almost all-eukaryotic plants contain well-developed chloroplasts. The color of plastids depends upon the dominance of specific pigments. Chloroplasts are bounded by two-unit membranes within which liquid is found, it is called stroma or stromatic fluid. Suspended within the stroma there are a number of highly organized membranous structures called grana and inter-granal membranes. Cyanophycean cells are almost like Plastids.
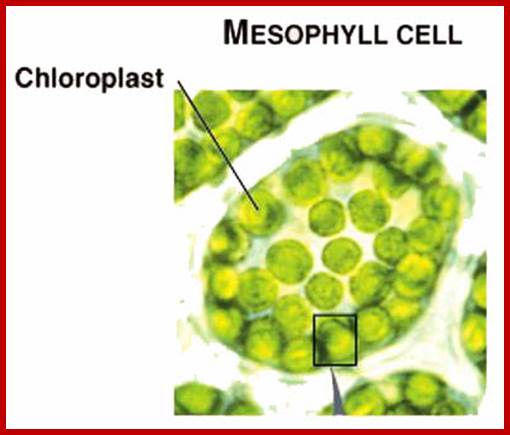
Mesophyll Chloroplastisds; Plant cell with plastids-www.hedgy.com; http://www.microscopy-uk.org.uk/www.tutorvista.com
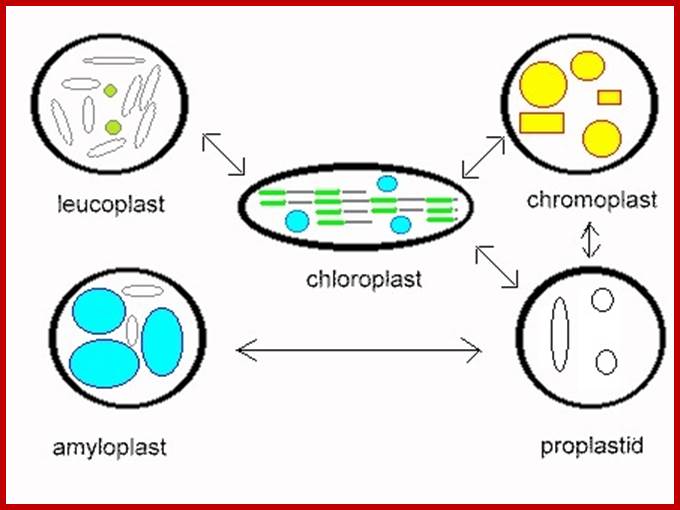
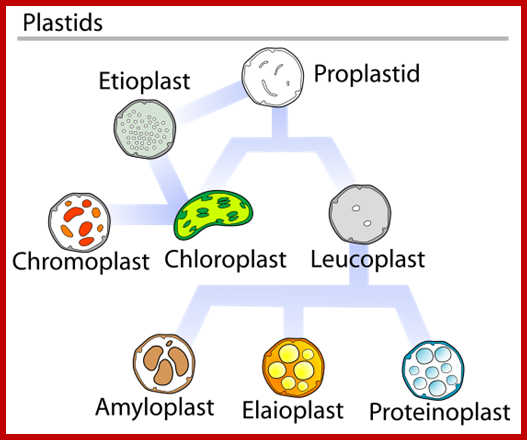
https://commons.wikimedia.org/
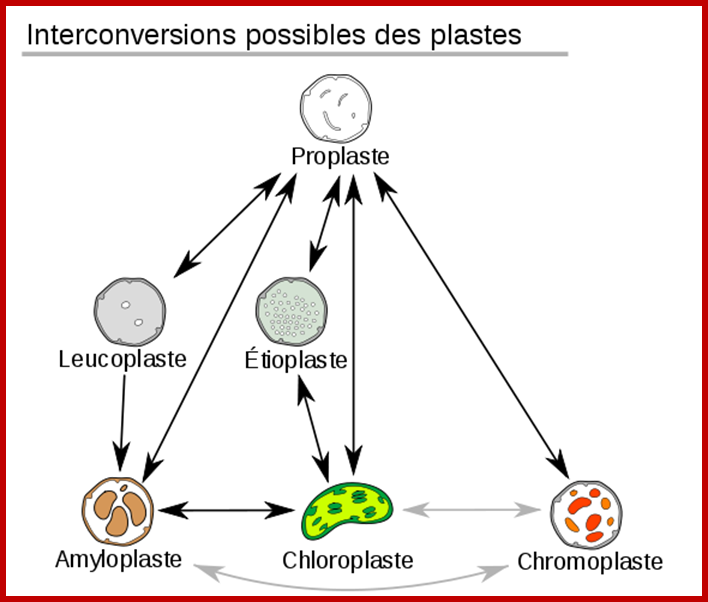
Plastid conversions; https://commons.wikimedia.org
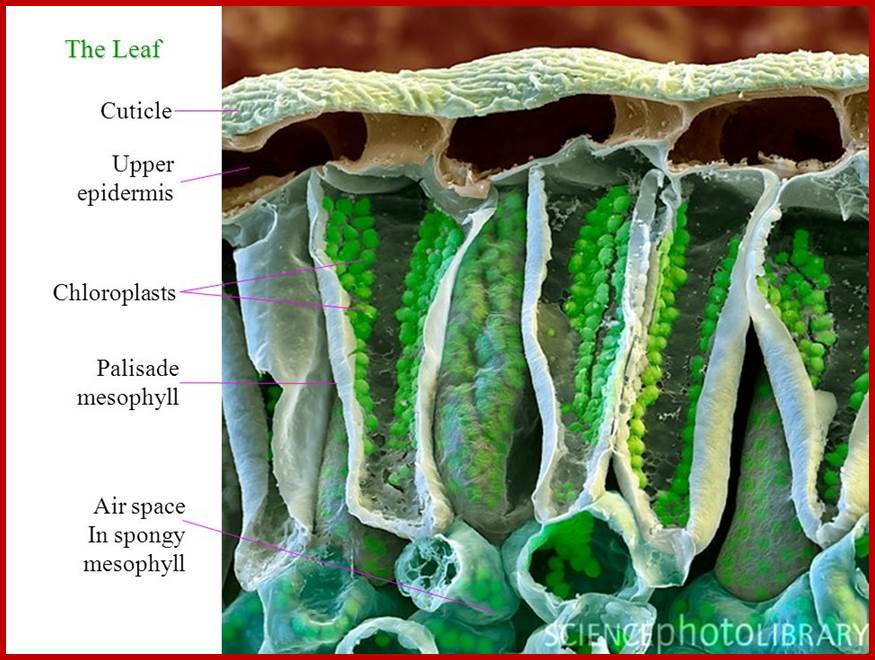
http://slideplayer.com/

Cell with central vacuole and peripheral plastids; www.s10.lite.msu.edu
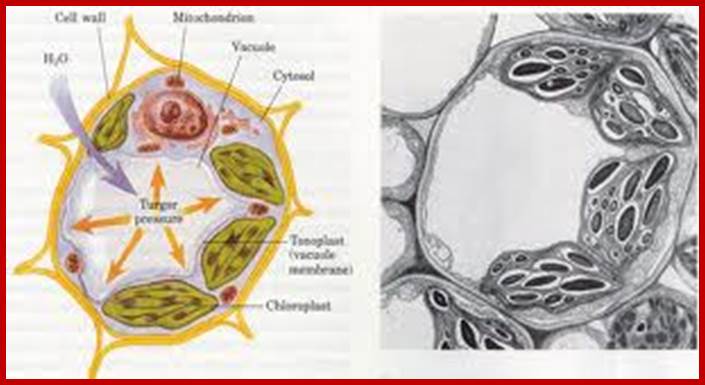
Central Vacuole in plant cells; www.xarquon.jcu.cz; www.quia.com
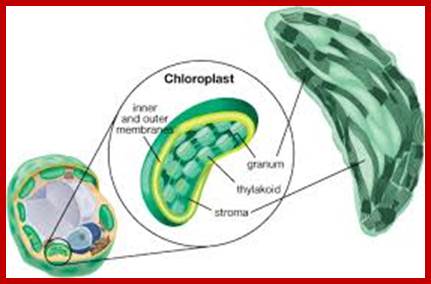
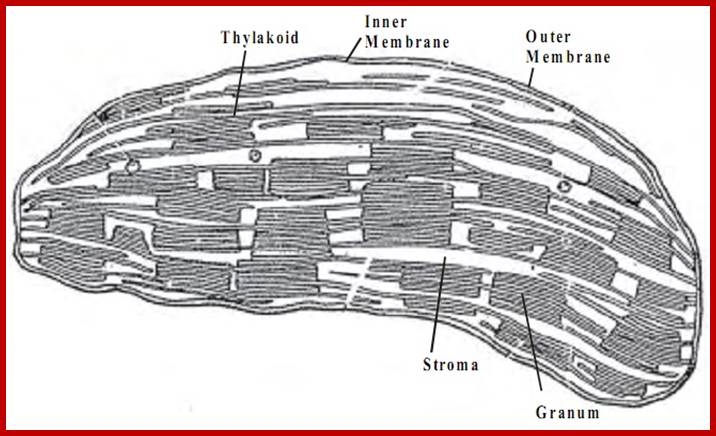
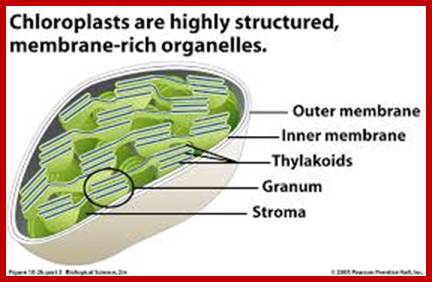
Chloroplast contain enclosed in outer and inner membranes. Inside the chloroplast is filled with fluid with the required metabolites and proteins and highly organized membrane sacs called grana and stroma and chloroplast DNA and RNA and ribosomes. Each granum is made up of 20-60 pigment/protein containing membranous sacs called Thylakoids organized one above the other; in the form of a stack. Besides granal structures, stroma also contains all the enzymes necessary for carbon fixation, protein metabolism, nucleic acid metabolism, fatty acid synthesis etc. Even certain phytohormones like Gibberellic acid (GA) and Abscisic Acids (ABA) are synthesized within chloroplasts. Chloroplast membranes also contain unique pigments called Phytochromes. The presence of 70s ribosomes and nucleic acids like DNA and RNAs (the genetic material) in chloroplasts provide information for its biogenesis, functions and inheritance of plastids. However, nuclear coded gene products are also required for the structural organization and functional efficiency of the organelle. That is why plastids do not enjoy complete autonomy, but functions as semi-autonomous organelles.
Chloroplast with granal and intergranular structures; www.tutorsglobe.com
Monocot bundle sheath Chloroplasts contain only stromal lamellae; www.tolweb.org
www.uic.edu

Graphics of granal structure; www.nature-education.org
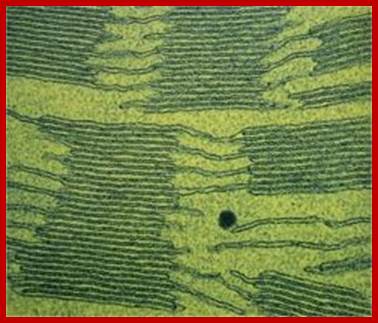
Granal Thylakoids and intergranal thylakoid (stromal) membranes; www.royalsociety.org
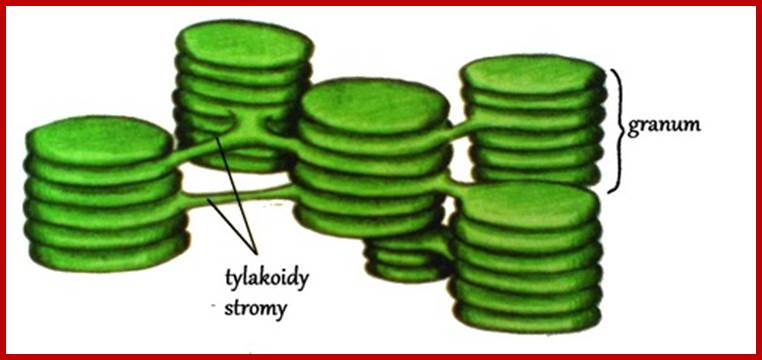
www.moja-biologia.blogspot.com
THYLAKOIDS:
Thylakoid is a circular flat membranous sac and such structures are arranged one above the other similar to a stack of carom coins. Such a stack of thylakoids is called Granum. However some of the thylakoid membranes found in a granum are continuous and they are in contact with other granal structures. Such lamellae are called intergranal lamellae or stromal lamellae.

Granum structure The prevailing model for granal structure is a stack of granal thylakoids linked by helical stromal thylakoids that wrap around the grana stacks and form large sheets that connect different grana; http://www.digplanet.com/;www.education-portal.com
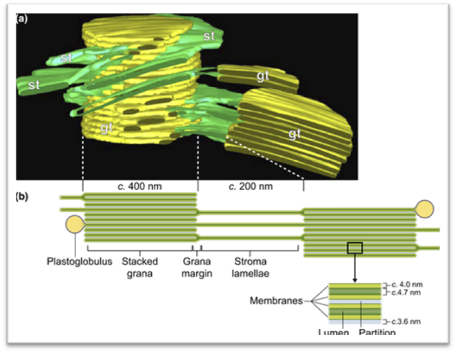
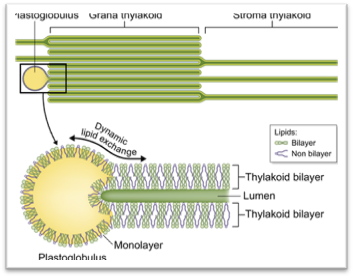
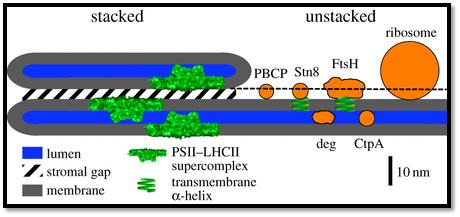
Ultrastructure of the thylakoid membrane system. (a) 3D reconstruction of the thylakoid architecture derived from electron tomography (ET) data (Austin & Staehelin, 2011; reprinted with permission from the American Society of Plant Biologists). gt, stacked grana thylakoid membranes; st, unstacked stroma lamellae. Openings in the grana cylinder represent membrane bridges called ‘frets' that connect stacked with unstacked membranes. (b) In-scale model of a thylakoid membrane cross section derived from (Kirchhoff et al., 2011; Herbst ova et al., 2012). The grana diameter can vary between 350 and 600 nm. The model reveals the strict stacking of membranes in the grana area. The diameter of plastoglobuli is typically between 45 and 60 nm but can vary under certain conditions. https://nph.onlinelibrary.wiley.com/
Open in figure viewerPowerPoint
Hypothesized role of plastoglobuli to maintain a constant lipid: protein ratio in thylakoid membranes. The upper panel cartoons part of the thylakoid membrane. The lower panel gives a model of plastoglobuli and its connection to the thylakoid membrane. About 50% of thylakoid membrane lipids are non-bilayer monogalactosyldiacylglycerol (MGDG) (lipid in purple) that have a small headgroup and a bulky fatty acid part leading to an overall conical contour. By contrast, bilayer forming lipids (green) adopt an overall cylindrical shape. Plastoglobuli are surrounded by a lipid monolayer that is physically connected to the outer leaflet of the thylakoid membrane bilayer. Therefore, it is likely that lipids can be exchanged between the plastoglobuli monolayer and the outer leaflet of the thylakoid membrane. Under conditions where the lipid : protein ratio in thylakoid membranes changes, the lipid pool in plastoglobuli can correct the altered lipid : protein ratio by either adding to or taking up lipids from thylakoid membranes. In chloroplasts, the lipophilic plastoglobuli lumen (yellow area) mainly contain prenylquinones, triacylglycerol and c. 30 different proteins involved mainly in isoprenoid and neutral lipid metabolism (Van Wijk & Kessler, 2017).
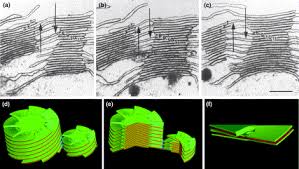
The complex structure of higher plant chloroplasts has fascinated researchers for many years. Although the spatial relationship between granum and stroma thylakoids has been known for more than 20 years, most textbooks and research papers continue to include erroneous 3D models and simplified schemes. Here we present a simple computer model, based on electron micrographs from serial section of granum–stroma assemblies, showing the striking 3D structure of the stroma membrane wound around the granum. This model also provides an insight into some previously unknown functions of this intriguing multilamellar membrane system. However, many areas, such as self-assembly, structural flexibility and evolutionary niche, still remain to be explored.www.cell.com
Closed membranes are thylakoid membranes and extended membranes at the top and bottom are stromal lamellae; they contain photosystem-I only; www.en.citizendium.org
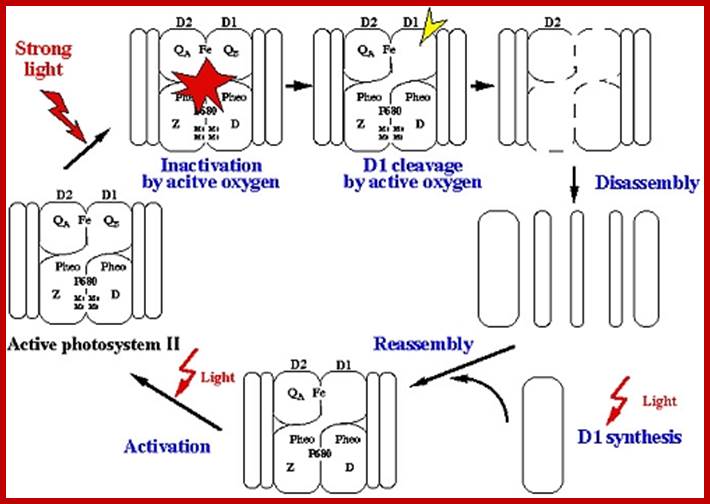
Structural model of the thylakoid membrane and components involved in PSII repair: The model represents the structural relationship between thylakoid membrane features and the sizes of proteins. The sizes and contours of the Deg and FtsH proteases were adapted from [50,51]. For PBCP, STN8 and CtpA, the sizes were calculated from their relative molecular mass (RMM). The RMM of a protein is proportional to its volume. Assuming a spherical protein contour, the relative change in diameter for protein 1 (dprotein1) to protein 2 (dprotein2) can be derived from RMM by solving the following equation for dprotein1: dprotein1/dprotein2 = (RMMprotein1)1/3/(RMMprotein2)1/3. The absolute diameter for protein 1 can be calculated if the relationship between d and RMM is known for a reference protein (protein 2). PC was selected as a reference protein. Its RMM is 10.5 kDa and its dimensions are 3 × 3 × 4 nm [52]. For the calculations, a mean diameter of 3.5 nm was used. Based on this number, the diameters of PBCP, STN8 and CtpA were calculated: PBCP: 32 kDa [40] ≥ approximately 5.1 nm; STN8 kinase; 56 kDa, extrinsic part 40.5 kDa ≥ approximately 5.5 nm; CtpA, 43 kDa [44] ≥ approximately 5.6 nm. (Online version in colour.); http://rstb.royalsocietypublishing.org/content

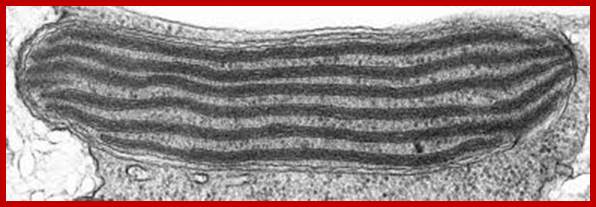
The electron micrograph on the left (courtesy of Kenneth R. Miller) shows the inner surface of a thylakoid membrane. Each particle may represent one photosystem II complex. In the functioning chloroplast, these particles may not be as highly ordered as seen here.; www.users.rcn.com

Thylakoid membrane surface and fractured surface view;www.visualphotos.com
www.pubs.rsc.org
Structurally, as shown in the figure, the surface area is the core of the thylakoid membrane in which a number of large globular shaped structures are located in such a way they span the entire cross section of the membrane and also a part of them protrude out at both the surfaces. Such larger particulates are called photosynthetic units II or photo system II , often they are called Quantasomes. In its latest form, the model suggests a bipartite structure consisting of a cylindrical granum body, made of discs piled on top of each other, around which the stroma lamellae are wound as right-handed helices. The grana are connected to each other solely via the stroma lamella helices, which are tilted at an angle of between 10 ° and 25 ° with respect to the grana stacks (Mustárdyet al., 2008; Daum et al., 2010; Austin and Staehelin, 2011) and make multiple contacts with successive layers in the grana through slits located in the rims of the stacked discs.

In this model granal stacks wound around by stromal lamellae in right-handed helical fashion; the granal discs are connected by a narrow membrane protrusions.
Thylakoid grana lamellae represent flattened sacs with diameter of ∼0.5–0.8 μm. The membrane thickness does not exceed 100 Å, and the maximal size of a protein complex embedded in it is ∼100–150 Å. Thus, it is a reasonable approximation to consider the lipid bilayer as a flat, two-dimensional surface. Photosystem II granules associated with photosystem I are mostly found in stacked thylakoid membranes, but stromal lamellae contain mostly Photosystem I. Such spatial segregation is called spatial separation of photosystems is called lateral segregation. Scientists have assumed that the photosystems carry negative charges of −1.6 × 10−18 C (PSI) and −1.2 × 10−18 C (PSII) and can move within a two-dimensional plaquette 0.6 × 0.6 μm2 (which is approximately four times the granal vesicle size) with periodic boundary conditions representing the thylakoid membrane. The density of particles in thylakoid membranes is about 2000-3000 per um^2. The ratio between PSII and PSI is about 7:3. And the particles carry negative charges and they can move in the membranes. The particles in lipid membrane cluster and segregate frequently. http://www.sciencedirect.com/
The membrane surface also contains a number of smaller particles. Some of them are photo system I structural units. The others are little bigger particles. They also contain Cyt f/b6 protein complexes in thylakoid membrane exposed to stromatic fluid. The enzyme complexes like ATP synthetase (F1) and RUBP carboxylase are located on this surface. In fact, a part of this complex of enzymes is buried in the membrane. In addition, there are some more protein complexes like ferredoxin reducing protein, NADP reductases and other electron transporting protein complexes within the thylakoid membranes. Most of these components are vectorially organized and moreover the above said particles show lateral movement within the dynamic fluid. The thylakoid membranous sac is filled with a fluid which is mostly acidic when chloroplasts are active.
The pigment-protein complexes within the membrane interact via Coulomb interactions (screened in the presence of cations), van der Waals (VDW) forces, dipole-dipole interactions, and lipid-induced protein-protein attraction.
The intergranal lamellae contain mostly PSI system and its associated components. The near absence of large PS II particles is a distinct feature of the stromal lamellae or intergranal lamellae. The presence of granular structures was first observed by Park and his associated members and such structures were then called as Quantasomes. But granal lamellae contain mostly PS II system. At the lateral ends of granal lamelle one finds both. And stromal lamellae are found coiled around granal structures
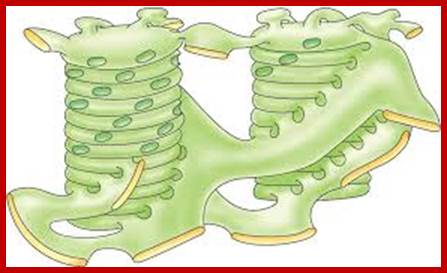
Thylakoid circular plates extended into stromal lamellae
There are two types of
thylakoids—granal thylakoids, which are arranged in grana, and stromal thylakoids,
which are in contact with the stroma. Granal thylakoids are pancake-shaped
circular disks about 300–600 nanometers in diameter. Stromal thylakoids are helicoid sheets that spiral around grana.
The flat tops and bottoms of granal thylakoids contain only the
relatively flat photosystem
II protein
complex. This allows them to stack tightly, forming grana with many layers of
tightly appressed membrane, called granal membrane, increasing stability
and surface area for
light capture.
In contrast, photosystem-I and ATP synthase are large protein complexes which jut out into the stroma. They can't fit in the appressed granal membranes, and so are found in the stromal thylakoid membrane—the edges of the granal thylakoid disks and the stromal thylakoids. These large protein complexes may act as spacers between the sheets of stromal thylakoids. Digplanet.com
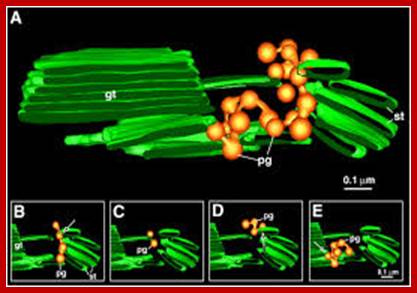
Plastoglobules; www.plantcell.org
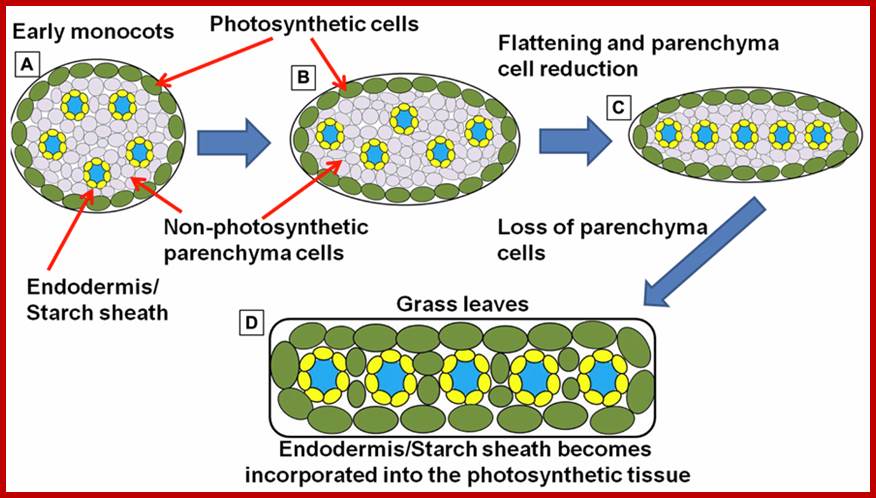
Simplified schematic representation of cross sections through monocot “leaf blades” along the evolutionary trajectory toward the grasses. (A)Simplified model of leaf structure in the early monocots (note: the early monocot leaves are depicted as radial structures in order to simplify the concepts presented). The vasculature encased in endodermal starch sheath tissue is separated from the outer photosynthetic layer by non-photosynthetic parenchyma cells. (B) The leaf structure begins to flatten and compress the vascular cores toward the center of the leaf, leading to a parallel vein patterning seen in (C). In grasses and sedges (D) the non-photosynthetic parenchyma cells are reduced or completely absent, bringing the outer photosynthetic layers in contact with the endodermis/starch sheath layer that surrounds the vasculature (model simplified and extrapolated form Arber, 1918).;http://www.frontiersin.org/
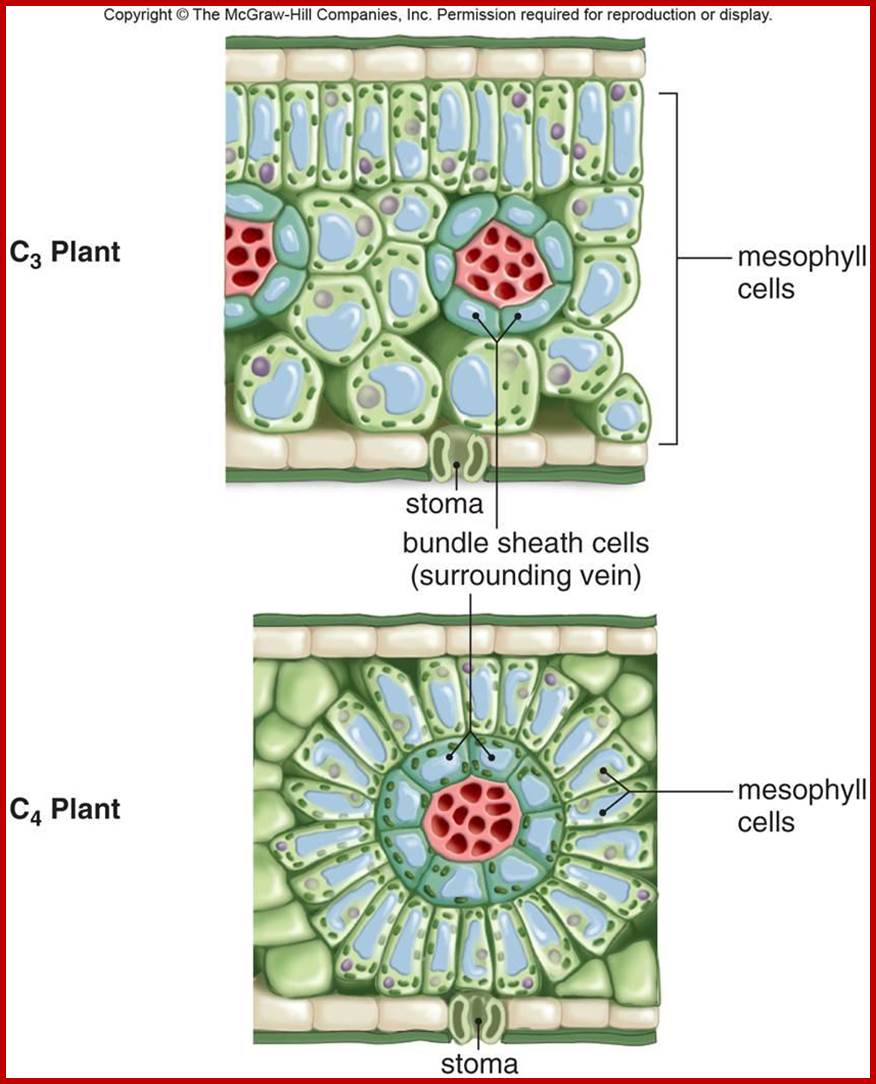
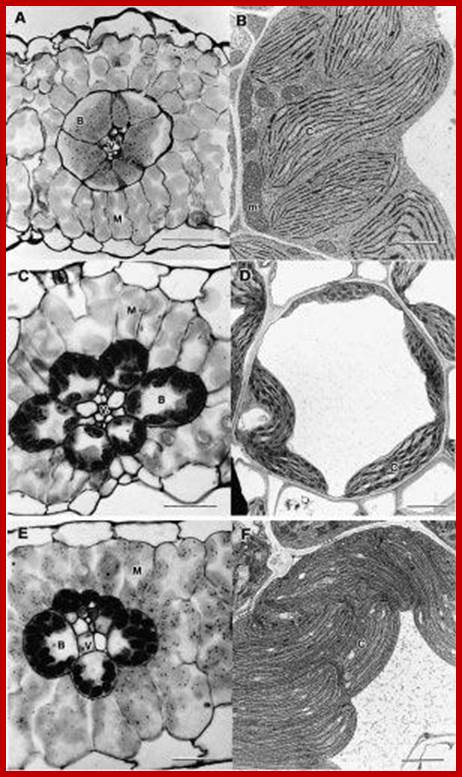
Not all chloroplasts show the structures as described above. Tropical grass members contain leaves where vascular bundles in leaves (veins) are surrounded by a distinct layer of cells called bundle sheath cells. Both mesophyll cells and bundle sheath cells contain chloroplasts, but with different structures and functions. While chloroplasts found in the mesophyll cells contain organized grana with intergranal lamellae; bundle sheath chloroplasts are totally lacking in granal membranes but contain only stromal lamellae with many large starch granules. Interestingly, the chloroplast membranes of mesophyll cells contain both PSI and PS II, but bundle sheath chloroplasts contain just PSI system and PS II is more or less absent. Another important feature that distinguishes mesophyll chloroplasts from bundle sheath chloroplasts is the presence of C4 pathway enzymes in the latter. Moreover, most of the chloroplasts found in bundle sheath cells are disposed towards their neighboring mesophyll cells and one can find a large number of protoplasmic connections between sheath cells and surrounding mesophyll cells. RubisCo found in large amounts in BS cell chloroplasts, whereas pyruvate phosphate di-kinase (PPDK) and photosystem II accumulate in mesophyll chloroplasts. The plants which possess such chloroplast fix carbon by Hatch and Slack pathway called C4 pathway hence called C4-plants. Ex. Tropical grass members like crab grass, sugarcane, Zea mays, etc. Even some dicots like Atriplex etc. also show C4 characters.
http://plantsinaction.science.uq.edu.au/
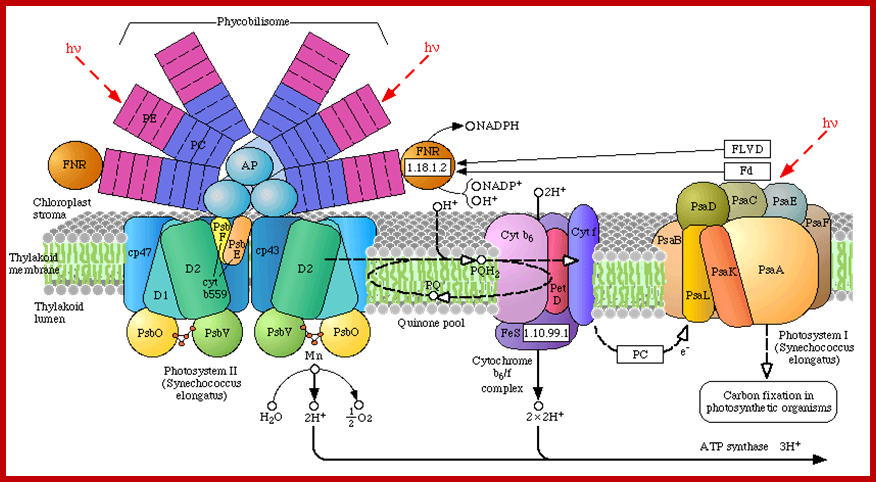
PHOTOSYSTEM I (PS I):
Recent techniques have been successfully used in isolating PS I and PS II systems into almost pure forms. The biochemical analysis of PS I reveal the presence of various plant pigments and different kinds of proteins. Each photosynthetic unit is made up of 250-300 chlorophyll molecules of which Chl.a is found in greater amounts (25 times) than Chl.b. There are just 40-50 molecules of Carotenoids in the entire complex. Along with the pigments a large number of light harvesting antenna proteins are present.
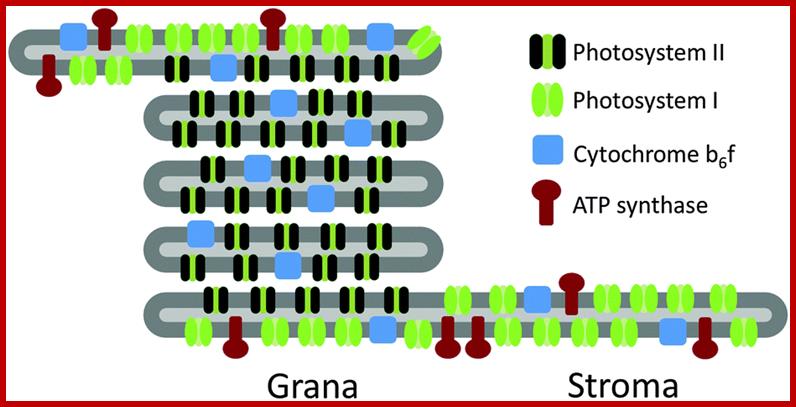
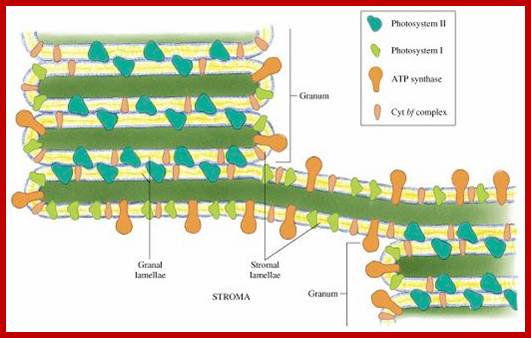
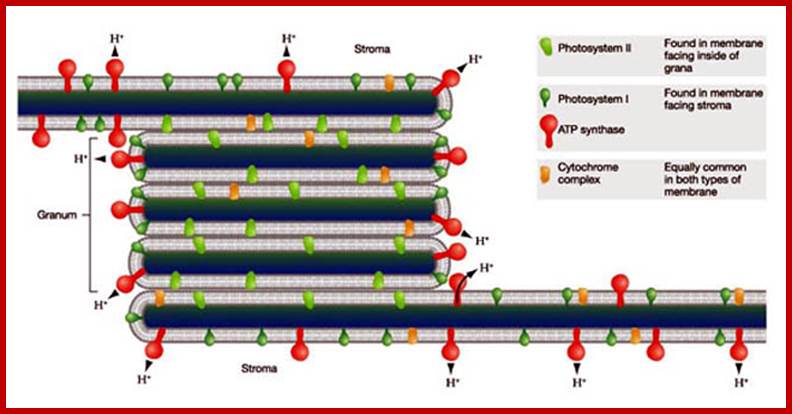
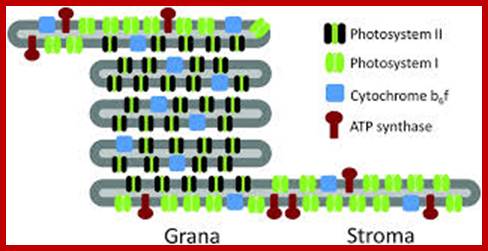
Difference between thylakoid and stromal lamella; Thylakoid membranes contain both PSI and PSII, but stromal lamellae contain only PSI; http://pubs.rsc.org/
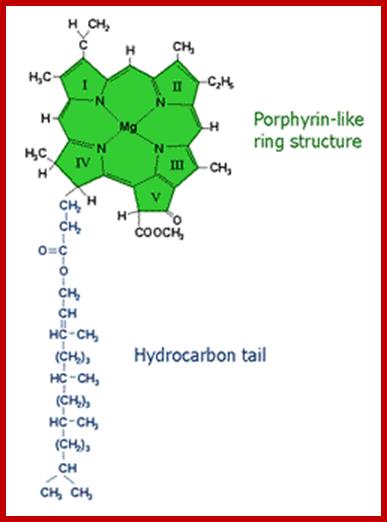
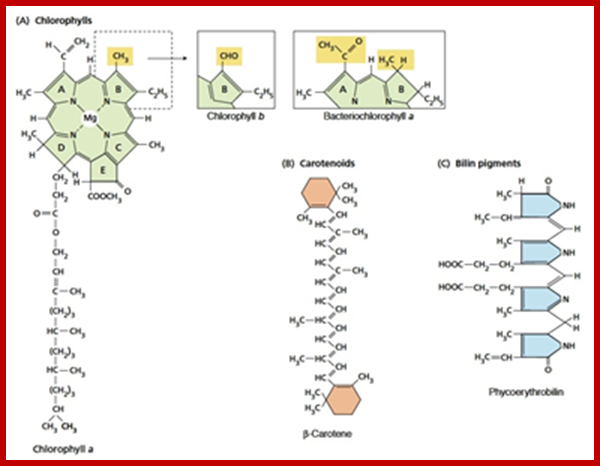
Molecular structure of some photosynthetic pigments; http://www.tankonyvtar.hu/
www.courses.ecampus.oregonstate.edu
www.en.wikipedia.org
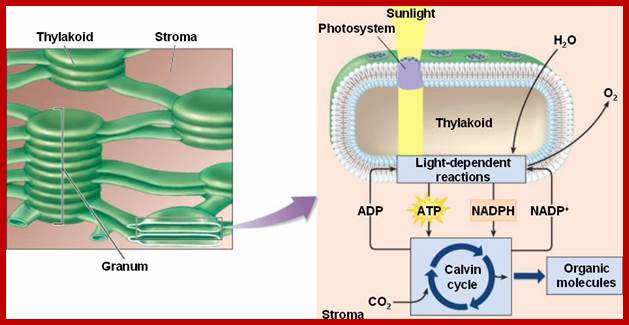
Thylakoids borne quantasomes capture sunlight energy at specific wave length and transfer it to cycle where energy released is used for generating NADPH2 and ATPs; http://bio1100.nicerweb.com/

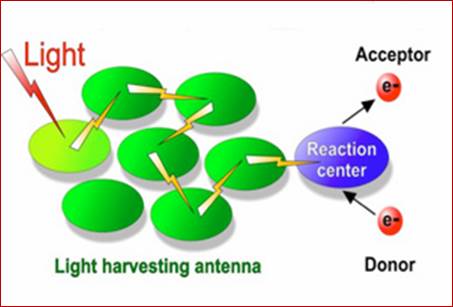
Organized pigments-antenna like; The antenna is a complex of chlorophyll pigments and its associated proteins; the antenna complex captures light in terms of quanta and the energy packets are transferred through special radiation free process called Foster Resonance Energy transfer called FRET, in which the electronic states of the sender and receiver are compatible. http://phys.org/news/
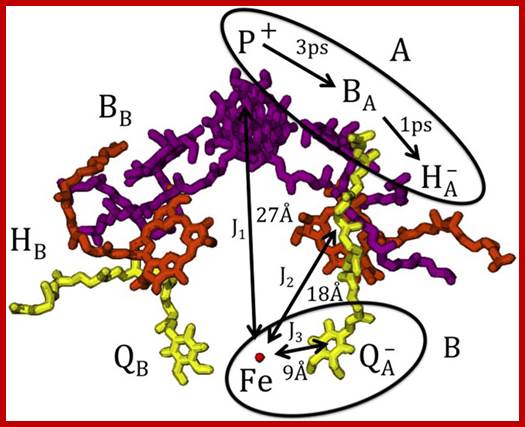
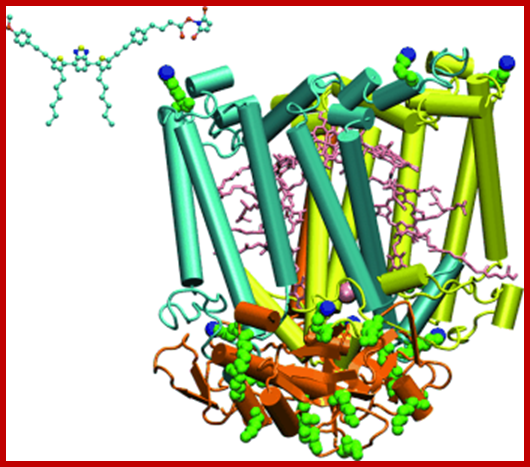
Shown are the
bacteriochlorophyll a dimer P and the bacteriochlorophyll a monomers BA and
BB (purple),
the bacteriopheophytin a molecules HA and
HB (orange), the ubiquinone molecules QA and
QB (yellow),
and the iron Fe (red). In RCs at room temperature with the ubiquinone removed
or reduced, the lifetime of the radical pair ![]() formed
on the active A-branch is
10-20 ns and singlet-triplet mixing is observed; http://www.nature.com/
formed
on the active A-branch is
10-20 ns and singlet-triplet mixing is observed; http://www.nature.com/
Chlorophyll Head, no tail; Giant water lilies in the Amazon have a very dramatic strategy to capture light for photosynthesis, by covering large surfaces of water. The leaves looking like huge circular trays may be as large as 6 feet in diameter; here is some excellent footage by David Attenborough, demonstrating this beautiful miracle of nature: http://islamforwest.org/
www.biology4isc.weebly.com; kvhs.nbed.nb.ca
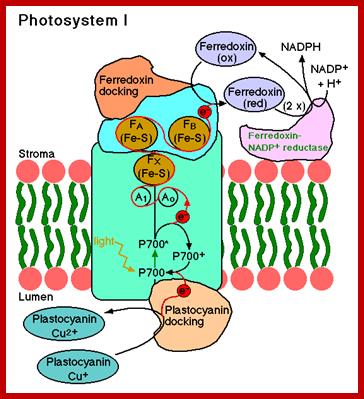
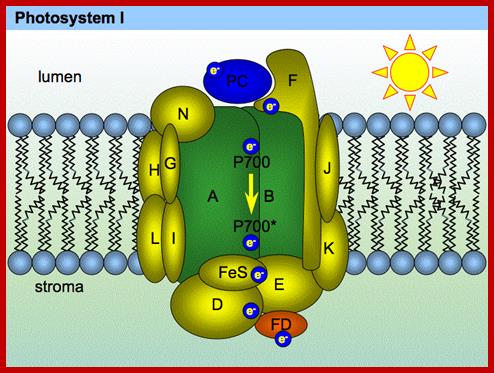
The mol. wt. of these proteins vary from 11 KD to 44 KD. The light harvesting proteins (LHPs) are associated with pigments in such a way; the captured light energy by chlorophyll-a pigments is conducted in a manner befitting a semiconductor. Among the LHPs, the core proteins are located in the dome shaped region where they are associated with two distinct Chl.a 700 molecules; which act as active or reaction centers. The other proteins called peripheral proteins are complexed with pigments they are located around the central dome. The peripheral proteins are capable of conducting photons channeled towards Chl.a 700, the active center of the PS I system.
In granal membranes, photosystem I particulate are also associated with ferrodoxin reducing Fe-S containing proteins (FRS), non-heme iron containing ferrodoxin proteins, FD dependent FD-NADP reductase and copper contained plastocyanin with protein complex. Some of the above mentioned protein complexes are as large as PS I systems and they are vectorially arranged in the membranes, so as to facilitate the electron flow from Chl.a 700 towards NADP. It is now certain that the ferrodoxin NADP reductase is topographically located towards the outer surface i.e. stromal side of the thylakoid membranes. However, the PS I units found in the intergranal lamellae contain most of the said pigments and protein complexes for light harvesting and photochemical reactions but they are not associated with FD NADP reductase. Instead, PS I am associated with Cyt.b6-Cyt.f are electron transport components, they are mobile from granal to intergranal membranes.
file:///C:/Users/user/Downloads/lec29-output%20(1).pdf;
www.plantphys.info
These photosynthetic units perform cyclic photophosphorylation reactions. In the granal membranes, photo system I in association with PS II is responsible for the reduction of NADP to NADPH2 liberation of oxygen and non-cyclic photo phosphorylation.
Top fig.; Cofactor arrangement in purple bacterial reaction centers (PBRCs) from R. sphaeroides (left), and in cyanobacterial photosystem I (PS I) from S. elongates; In PBRCs the primary electron donor is a dimeric bacteriochlorophyll-a (Bchl-a) species called P870. Upon light excitation of P870, an electron is transferred from P870* to HA (bacteriopheophytin-a) within ~3 ps. From HA¯, the electron is then transferred to QA in ~200 ps, and then to QB in microseconds. In PS I, following light excitation of the primary donor (P700), an electron is transferred to A0, a chlorophyll-a (Chl-a) acceptor, in ~4 ps. Stabilization of the charge-separated state is achieved by a second ET process; From A0¯ the electron is transferred to A1, a phylloquinone (PhQ) molecule in ~21 ps. http://www.phy-astr.gsu.edu/; Bottom Fig; Structure of Photosystem-I (PSI); www.nature.com
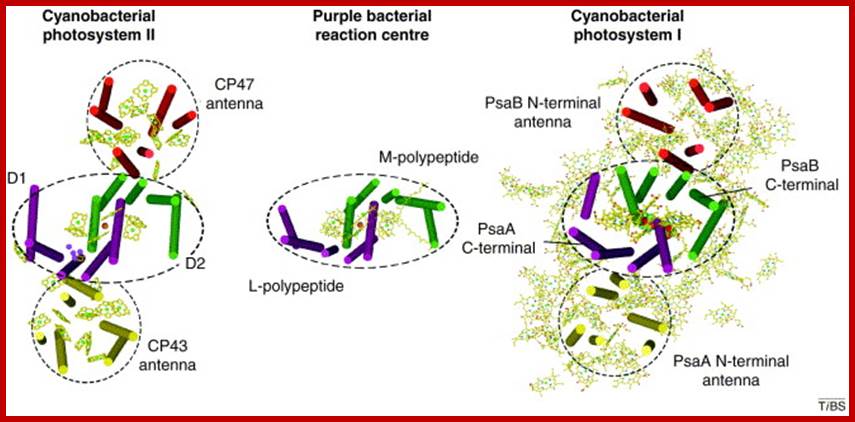
www.cell.com
Oxygenic photosynthesis is the principal producer of both oxygen and organic matter on Earth. The conversion of sunlight into chemical energy is driven by two multisubunit membrane protein complexes named photosystem I and II. We determined the crystal structure of the complete photosystem I (PSI) from a higher plant (Pisum sativum var. alaska) to 4.4 Å resolution. Its intricate structure shows 12 core subunits, 4 different light-harvesting membrane proteins (LHCI) assembled in a half-moon shape on one side of the core, 45 transmembrane helices, 167 chlorophylls, 3 Fe–S clusters and 2 phylloquinones. About 20 chlorophylls are positioned in strategic locations in the cleft between LHCI and the core. This structure provides a framework for exploration not only of energy and electron transfer but also of the evolutionary forces that shaped the photosynthetic apparatus of terrestrial plants after the divergence of chloroplasts from marine cyanobacteria one billion years ago. Adam Ben Shem, Nathan et al- http://www.nature.com/
Photo system II (PS II)
Photosystem II are larger photosynthetic units found in thylakoid membranes and but absent from intergranal membranes. They are also made up of pigments and protein complexes. The number of chlorophyll molecules found are 250-300, of which the number of Chl. A present is 7.5 times the number of Chl. B. That means the number of Chl. B molecules present in PS II are many times greater than Chl.B found in PS I system. The PS II also contains about 80-100 molecules of carotenoids. The protein molecules found in PS II vary in their mol. wt from 25 KD To 55 KD, of which the core proteins have a mol. wt. of 42-55 KD. This forms the central dome shaped structures which again possesses two unique chlorophyll molecules called Chl.A 680. They act as reaction centers. The peripheral complexes act as light harvesting structures. And most of these proteins are complex with various pigments.
Turning Plants into power houses; plant leaves are like solar photovoltaic solar panels.
http://news.wustl.edu/
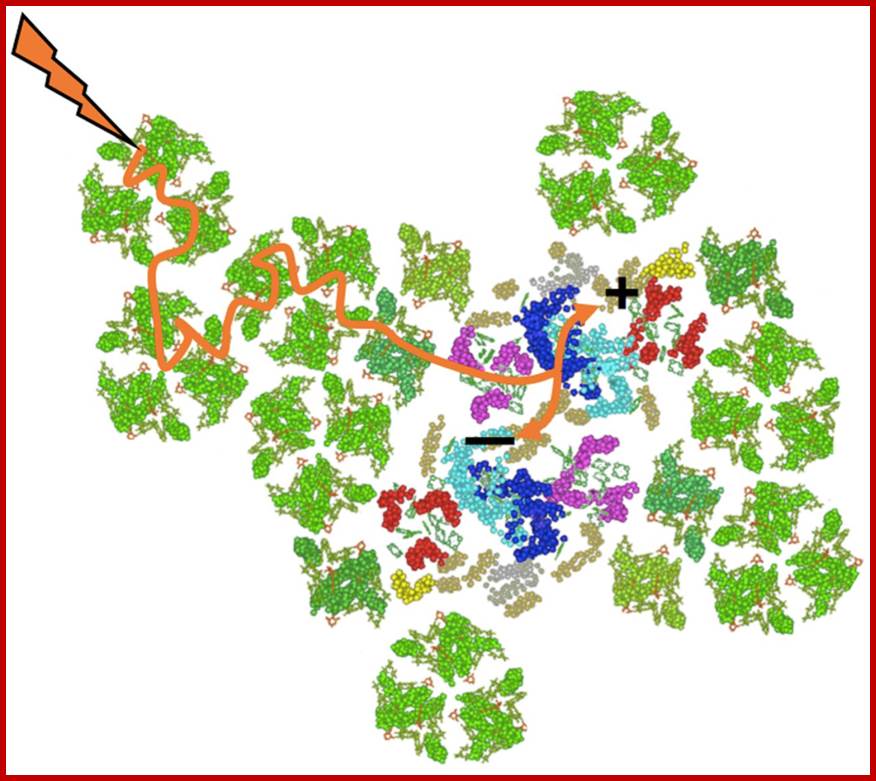
Principles of light harvesting from single photosynthetic complexes; G. S. Schlau-Cohen; http://rsfs.royalsocietypublishing.org/
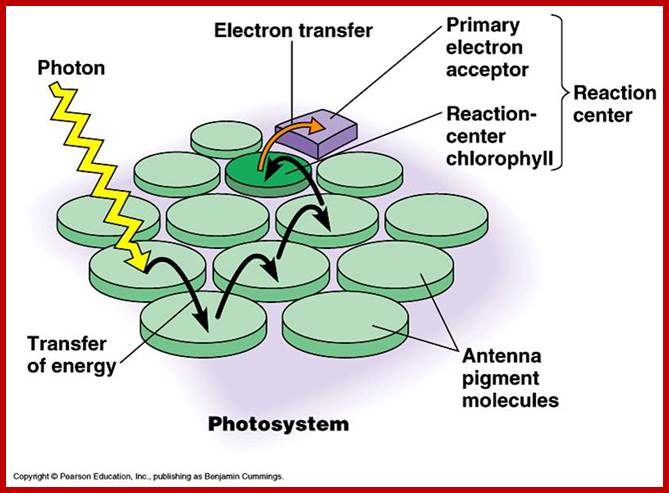
www.biology4isc.weebly.com; kvhs.nbed.nb.ca
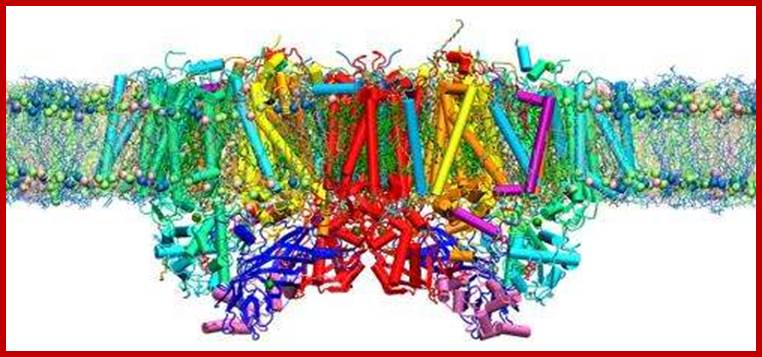 Computational
simulations of photosystem II inside a realistic, water-containing membrane
reveal the existence of previously hidden molecular pathways that play critical
roles in photosynthesis. Credit: American Chemical Society
Computational
simulations of photosystem II inside a realistic, water-containing membrane
reveal the existence of previously hidden molecular pathways that play critical
roles in photosynthesis. Credit: American Chemical Society
Read more at: http://phys.org/news/2013-12-exposing-secret-pathways-photosynthesis.html#jCp;
Exposing the secret pathways behind photosynthesis; http://phys.org/news/2013
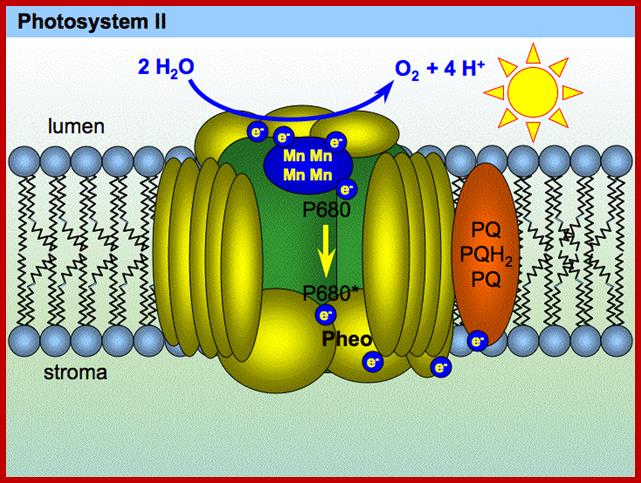
PSII; www.extremetech.com
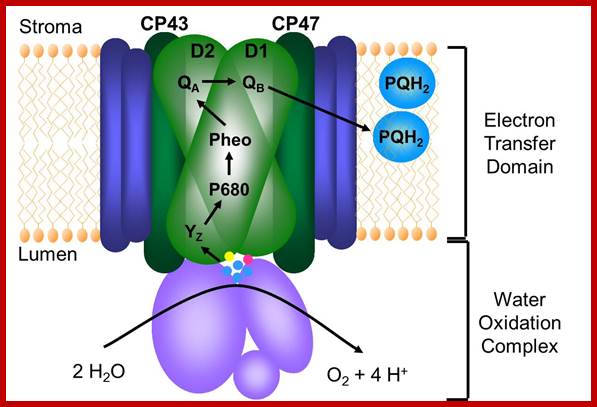
Photosystem II complex; Johanna; https://newunderthesunblog.wordpress.com
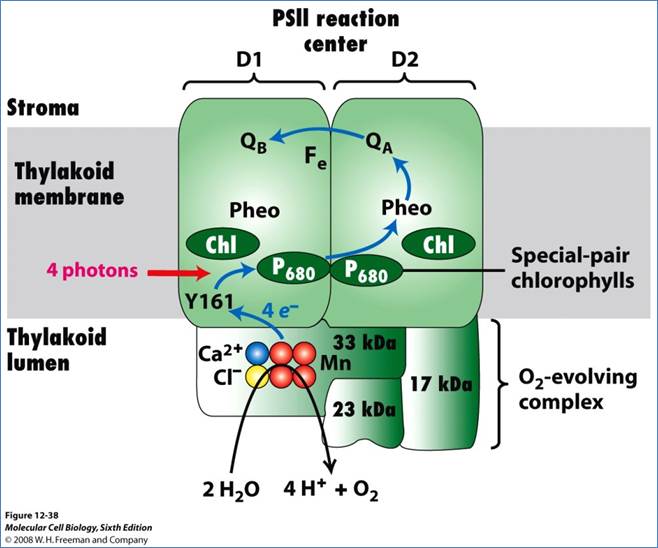
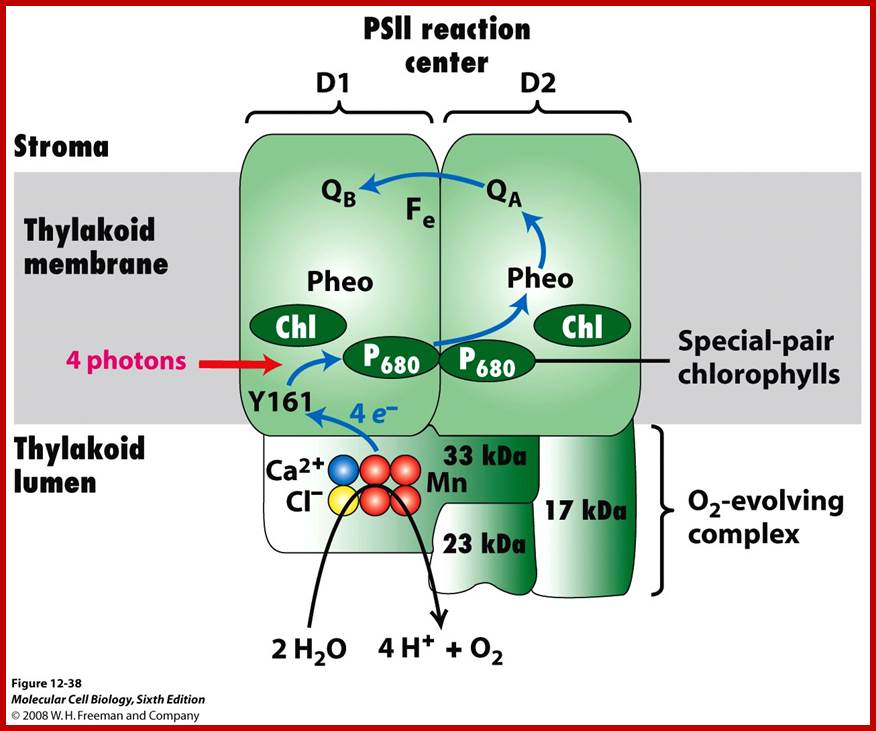
The above said photosynthetic units also possess another important protein called ‘Z’ protein, which is complexed with 4-6 Mn2+ molecules. The Z protein is highly hydrophobic. The other complexes which are associated with PS II are cyt.B559 and cyt.f reducing protein complex, pheophytin containing proteins and plastoquinones.
Recently, Kenneth Miller (1984) used techniques like deep etching, neutron diffraction and rotator shadowing to understand the structural organization of photosynthetic units in the chromatophores of Rhodopseudomonos viridis. In these bacterial cells, the photo synthetic units are organized within the chromatophores membrane as an array of regular crystal lattices. Each photosynthetic unit is 10 mm thick at the base and it has a central elevated dome of 7 mm thick. The central dome shaped complex acts as the photoreaction centre. This central dome in turn is surrounded by six peripheral units of 3 mm thickness each. The photoreaction centre is made up of special Chl.a molecules which are complexed with five LHPs of mol. Wt. 24-44 KD. The peripheral complexes also contain pigments associated with LHP of 11-16 KD sizes. Each of the photosynthetic units weighs about 423 KD. Now, it is speculated that the structural organization of photosynthetic units found in higher plants also have similar structural organization.
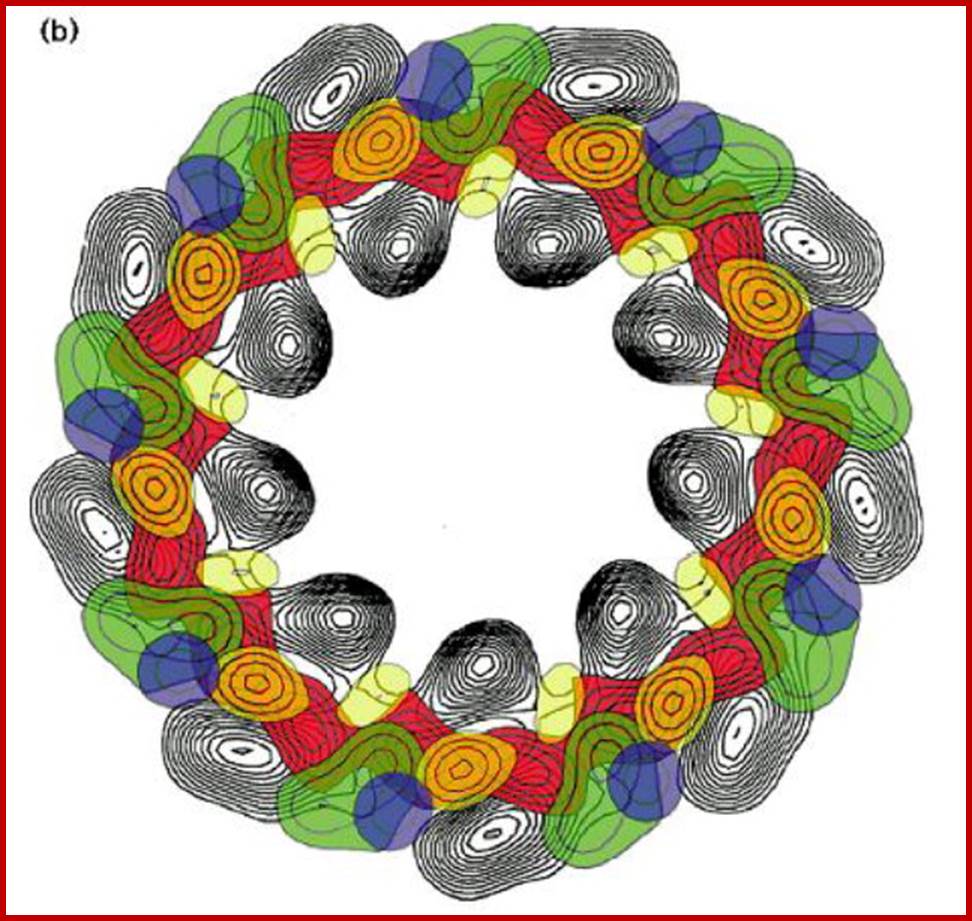
2-D of Light harvesting complex; Hugh Savage et al; Sciencedirect.com; www.cell.com
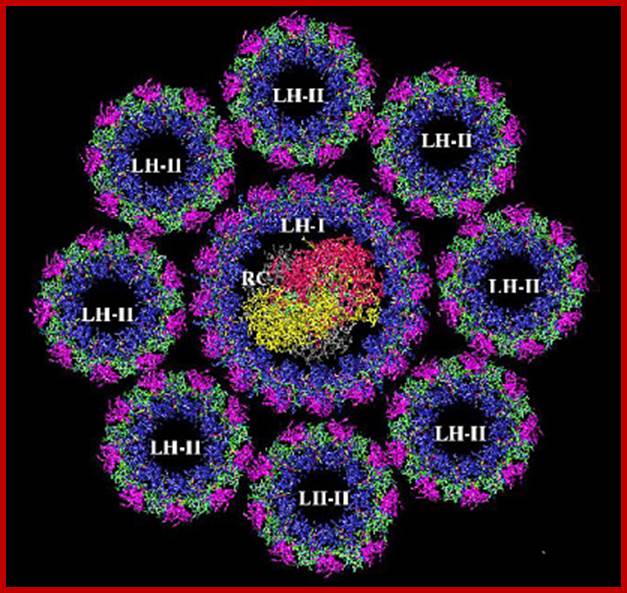
Organized LHI and LHII; www.chm.bris.ac.uk
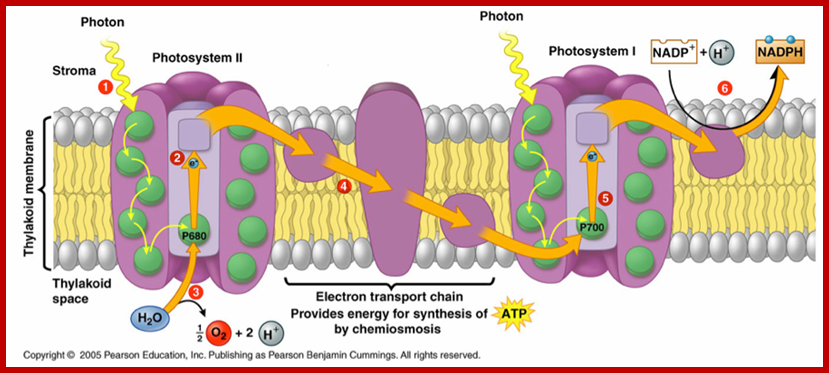
Photosystems PS1 and PSII; www.life.illinois.edu
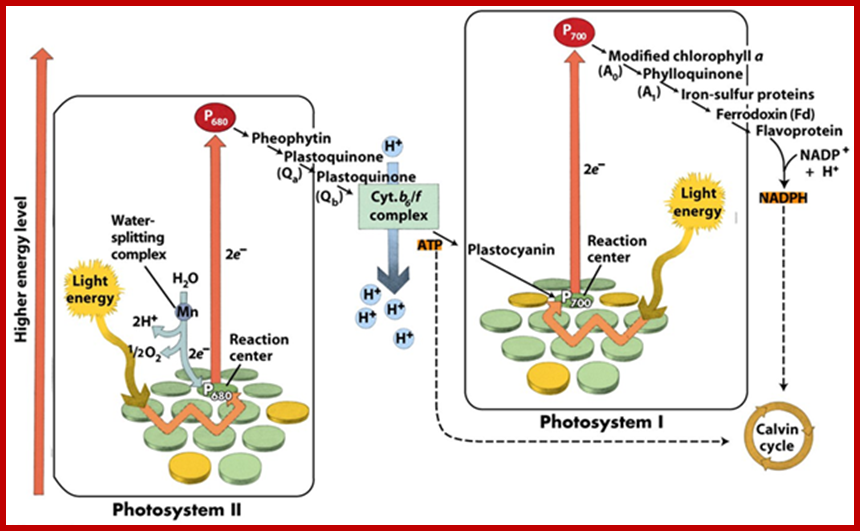
http://www.majordifferences.com/
Thomas M. Brennan; Basic Photosynthesis; http://www.photobiology.info/
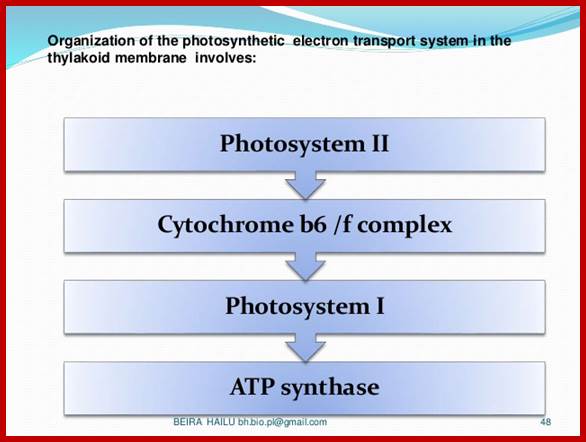
Thylakoid membrane contains both Ps-I and Ps-II system and ATP synthase
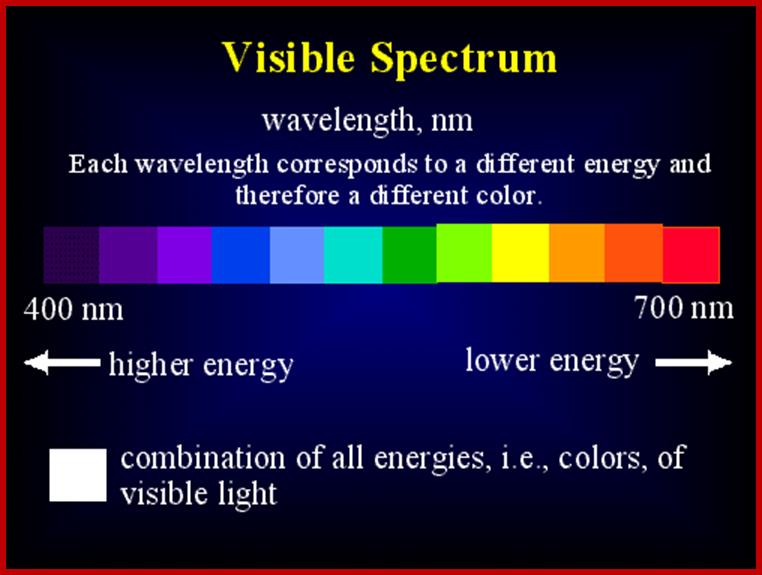
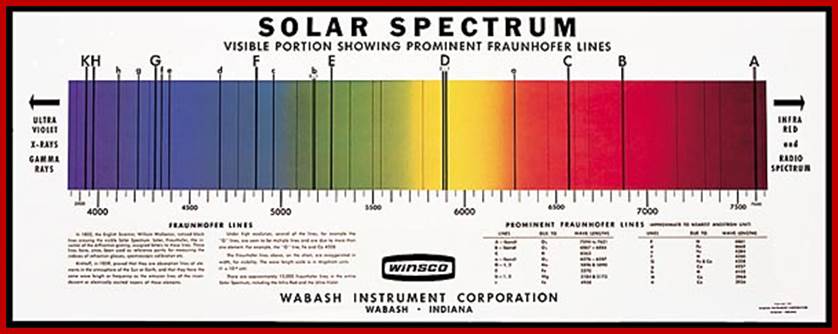
ABSORPTION SPECTRUM VS ACTION SPECTRUM:
When, chloroplasts in Toto are allowed to absorb white light at different wavelengths they exhibit maximum absorption (peak) at red, blue, green and yellow part of VIBGYOR spectrum and it is called absorption spectrum.
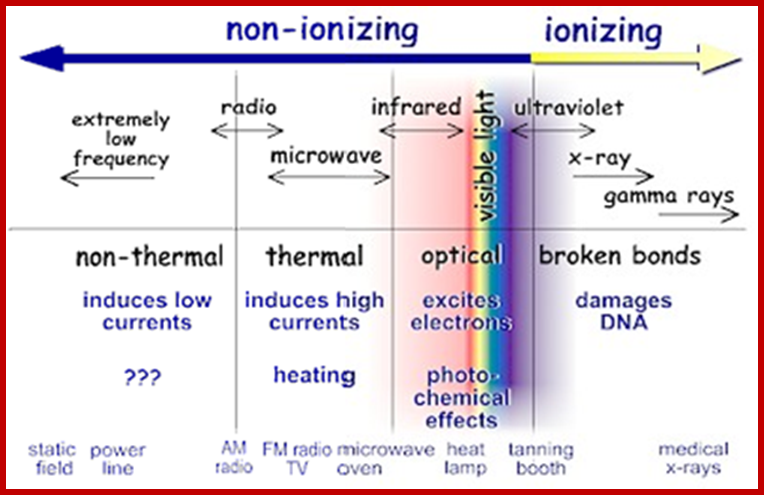
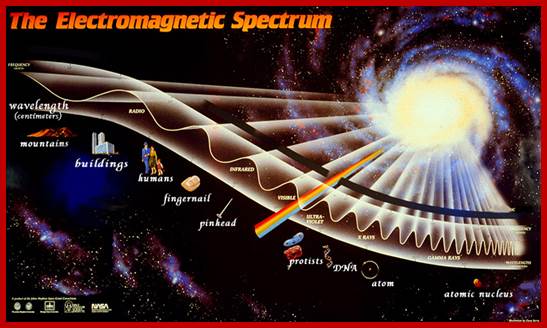
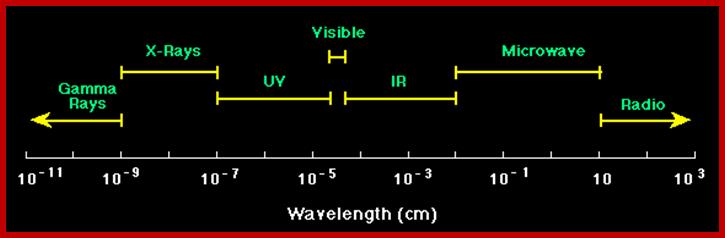
The diagram below shows the full electromagnetic spectrum and everything it contains. On the red side of the spectrum are low-energy electromagnetic waves, and on the blue side of the spectrum are high-energy electromagnetic waves; http://education-portal.com/
On the other hand, if specific pigments are allowed to absorb light at different wavelengths, chlorophyll shows maximum absorption at 435 mm and 670-680 nm. Similarly Chl. B shows absorption peak at 480 and 650 mm, but carotenoids show a broad absorption spectrum ranging from 420-524 nm.
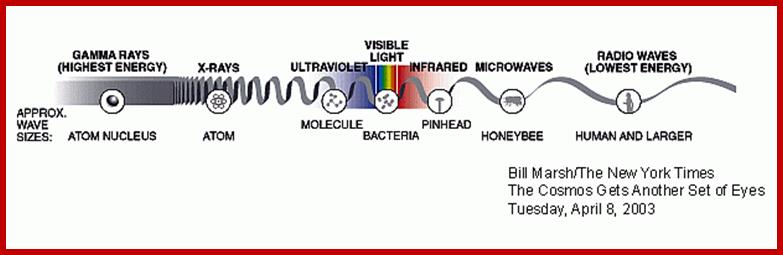
Photon as an unit, quantized with energy depending upon the wavelength.
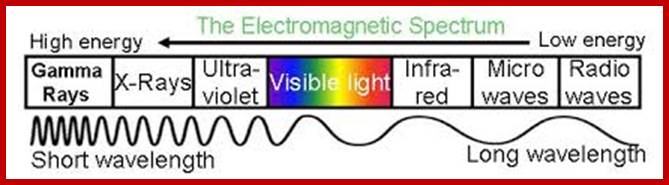
Electromagnetic spectrum; www.imagine.gsfc.nasa.gov; www.thesolarspark.co.uk


Antenna in tree leaves form to harvest energy; webclass.angelo.edu
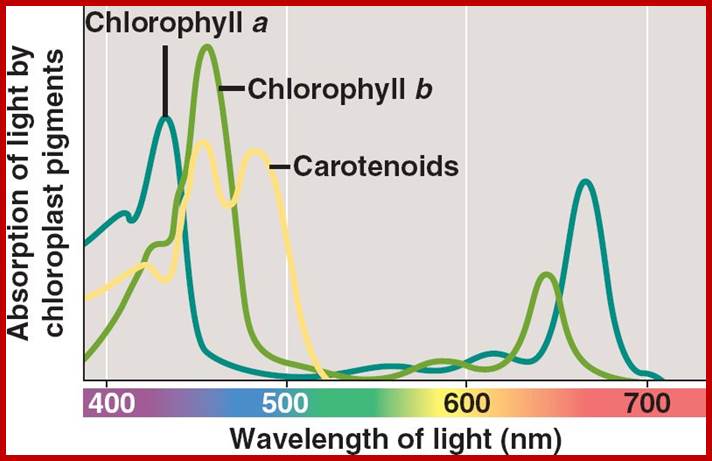
Absorption; http://users.rcn.com/jkimball; bio1903.nicerweb.com
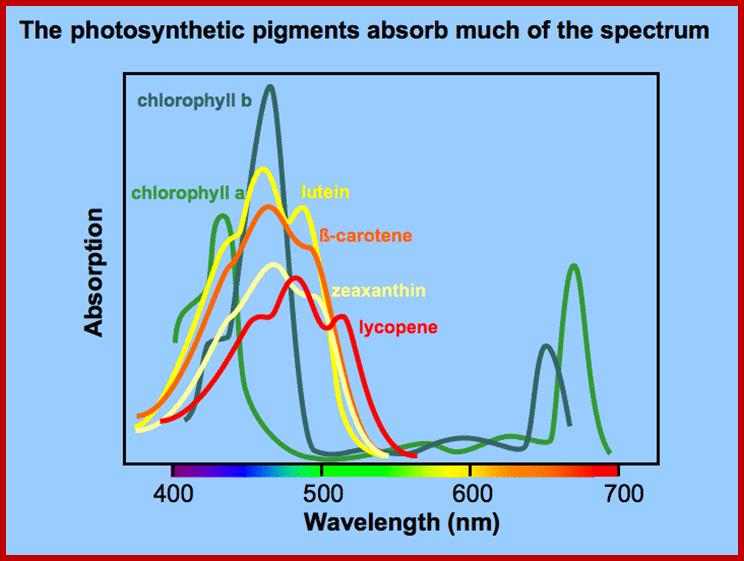
Wave-lenght of light absorbed by different pigment systems; http://plantphys.info/
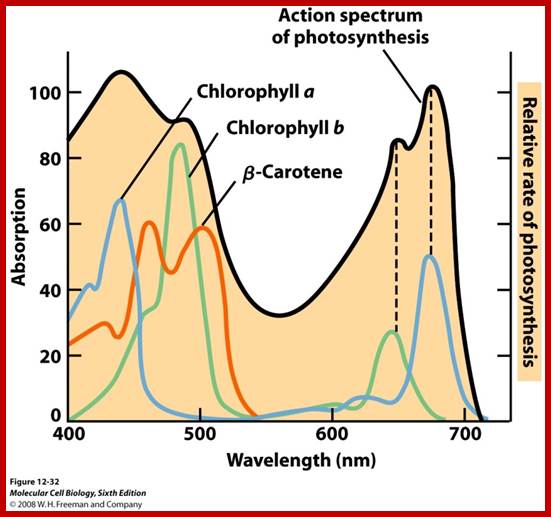
Absorption spectrum; http://www.bio.miami.edu/
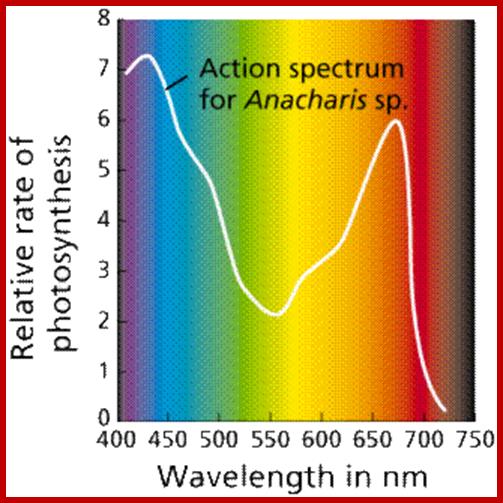
Action spectrum is the spectrum where action takes place in terms of activation of pigmants; ;www2.estrellamountain.edu
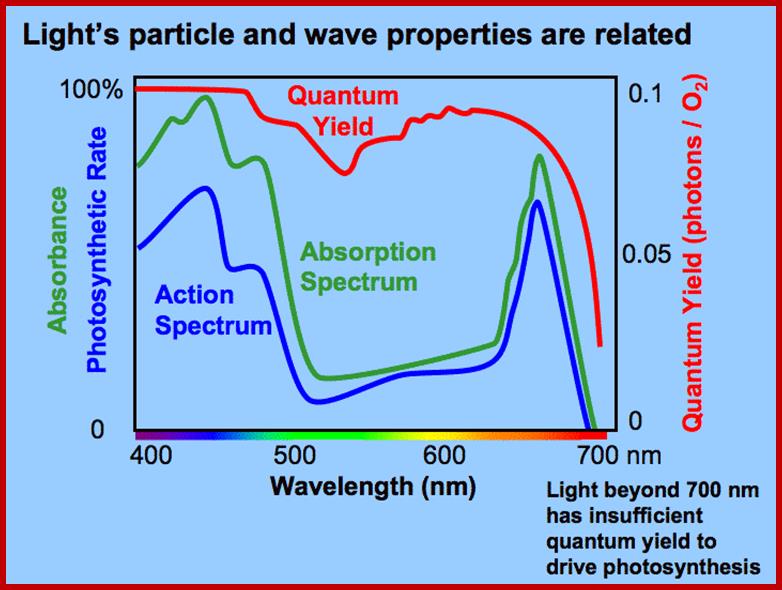 Absorption
spectrum and action spectrum; ;www.plantphys.info
Absorption
spectrum and action spectrum; ;www.plantphys.info
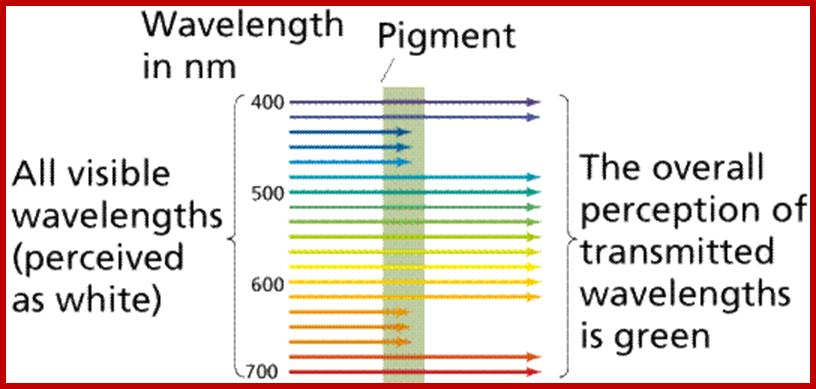
The part of light that is mainly responsible for photochemical reactions is called action spectrum. Though leaves absorb light at red, blue, green, orange and yellow regions, all are not used for photochemical reactions. It is possible to distinguish between the absorption spectrum and action spectrum by subjecting green leaves, unicellular algae or isolated chloroplasts to single beam of monochromatic light at a particular wavelength or a combination of wavelengths; and observe for the release of oxygen or carbon fixation into glucose. The part (wavelength) of light or a combination of light wavelengths that effectively initiates and sustains photosynthesis is considered as action spectrum. Studies in this regard have revealed that blue and red lights are the action spectrum. If blue or red light are alone used, though there is an initial reaction, the process of photosynthesis does not continue. For the sustained photosynthetic reactions both blue and red lights are required. Such an affect is called Emersion effect or Emersion’s Enhancing Effect. Furthermore, Emersion and others found that if the wavelengths of red light beyond 700 mm along with blue light are used photochemical reactions are inhibited. Such an effect is called Red drop effect. The above observations suggests though plants absorb light at various wavelengths of VIBGYOR spectrum but only blue and red light at particular wavelengths are effective in photosynthesis.
TRANSFER OF LIGHT ENERGY
From the analysis of absorption spectrum and action spectrum, as discussed above, it is clear that though plants absorb light at different wavelengths, the absorption bands at 435 and 680-700 are very important for they alone initiate photochemical reactions.
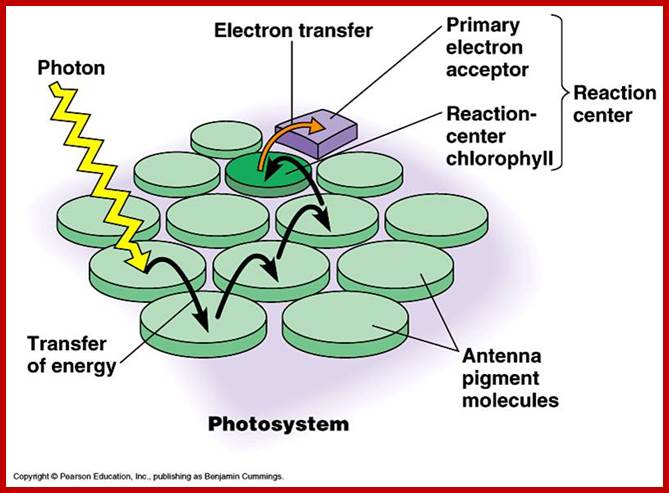
Nonetheless, the white light that is absorbed at other wavelengths is not wasted but used by photo systems. Photons absorbed at different but specific wavelengths by different pigments, transfer the light energy as quantized units from molecule to molecule through light harvesting proteins. The unit of light energy that is transferred is often referred to as excitons and the process by which excitons are transferred is resonance mechanism.
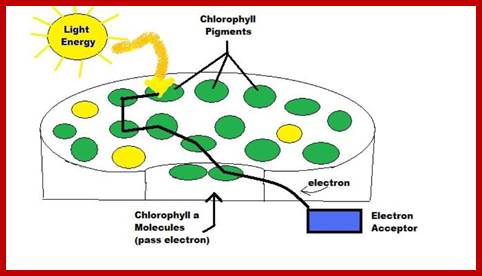
Absorbed light particles transferred from one pigment to the other by resonance, ultimately to primary electron acceptor; www.en.academic.ru
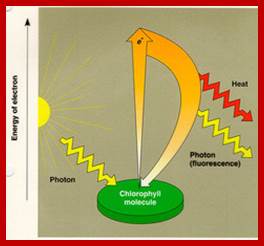
When an electron raised to higher energy state, it falls down at different levels., where energy is released, which can be used;www.quizlet.com
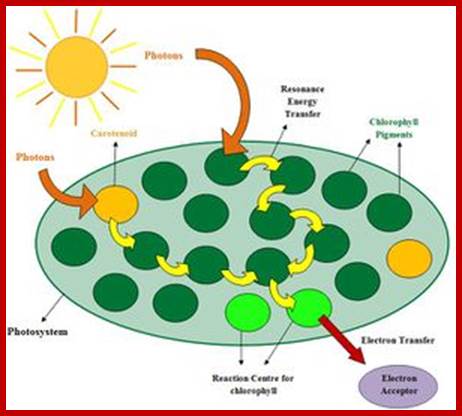
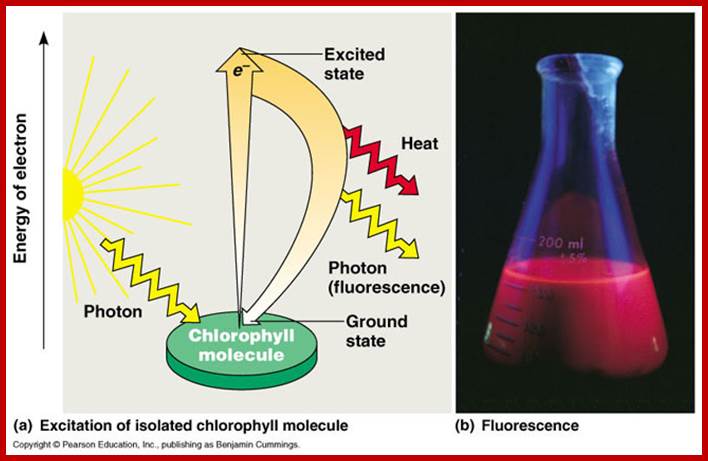
A simplified representation of chlorophyll’s role in capturing light energy of photons ‘units’ of light energy and converting it to electrical energy in the process of photosynthesis. https://microbewiki.kenyon.edu
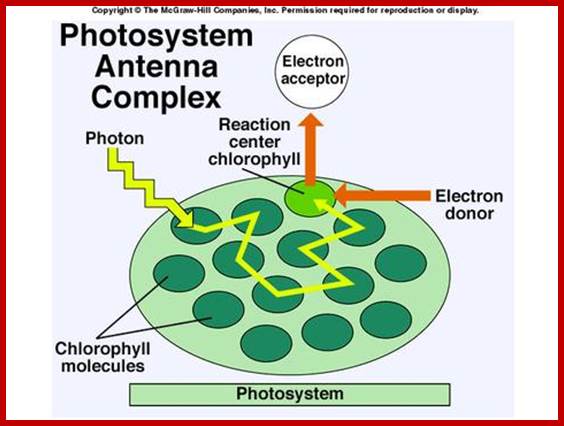
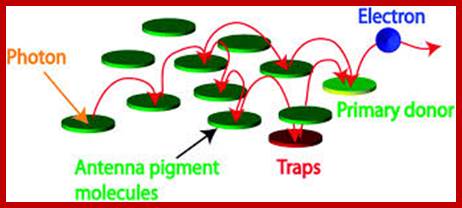
www.zoology.okstate.edu;www.deisenhofer.checknobel.com
The photons absorbed by carotenoids are transferred to Chl. B or Chl.A. Similarly, light energy absorbed by Chl. B is transferred to Chl.A. Again among the Chl.A molecules, light energy absorbed at 683 mm is transferred to Chl.A 680 or Chl.A 700. In this process, generally the pigments that absorb light energy at shorter wavelengths then they handed over by resonance to the pigments that absorb or capable of absorbing light at longer wavelengths. The light harvesting antenna proteins play an important role in transferring the excitons as semiconductors. Whatever may be wavelength of the light absorbed or whatever may be the pigments that absorbs light, ultimately the light energy in the form of excitons are channeled to photo reactive centers in the photosynthetic units. In PS II the light energy is ultimately conveyed to Chl.A 680. Similarly in PS I system, light energy is transferred to Chl. A 700. The transfer of energy from molecule to molecule is not cent percent. For example, the transfer of energy among the accessory pigments or accessory to primary pigments is 80-90%, but among Chl.A pigments it is 100%.
SOLAR ENERGY AND ITS PRIMARY EFFECT IN PHOTOSYNTHESIS:
All living organisms require chemical free energy for their biological activities. The ultimate source of this form of energy comes from the sun as solar electron magnetic radiations. Among all the living organisms, plants with the exception of fungi are alone capable of capturing, converting and conserving the solar energy in chemical bonds in organic molecules like glucose and its derivatives. The same is made available for other living organisms. That is why the food that we eat is considered as bottled sunshine. In fact, the flesh that develops in animals is derived from the green grass. And all the sources available for mankind in the form of fossil, fuel or bio fuel is nothing but photosynthetic capital.
Sun, for that matter any other living star in our universe, by its nuclear reactions liberates enormous amount of energy in the form of solar electromagnetic radiations. These radiations have a wide spectrum of which the visible i.e. perceptible to human eye is mainly responsible for initiating photochemical reactions in plants. The red and blue part of VIBGYOR spectrum is mostly utilized in the process of photosynthesis. Out of the total amount of solar energy that strikes the surfaces of Photosynthetically active plants parts, only a small portion of it is fixed in the form of chemical energy and the rest of it is reflected or lost as radiation energy. The total amount of solar energy that is fixed by the plants on this planet has been estimated to be 13 x 18 K.cals per year which is actually used to fixation of 160 to 175 billion tons of carbon per year, out of which nearly 130-135 billion tons of carbon is fixed by the plants living in oceans and the rest by land plants. Furthermore only 40-50% of it is tapped by human beings and other organisms, the rest is fixed as biomass for future use.
Where h=n Planck’s constant (6.624+10-27 erg sec.) = frequency of light waves per sec. C=velocity of light (2.998 x 1010 cms/sec.) and x = wavelength in centimeters. According to Einstein’s law of Photochemistry, in any photochemical act, one atom or one molecule can absorb only one photon at any given time and this brings about only one reaction at a time.
In order to relate the photochemical effect and the molecule that is affected by it, Avogadro’s molar values have been used to explain mass action i.e. E=NHV, where E is total energy found in mole quanta of light at any wavelength. Thus one Einstein of red light at 600 mm i.e. one mole of red light photons (6.2 x 1023 photons) possess 47.667 K. cals of energy. So different bands of VIBGYOR show different levels energy per quantum. Shorter the wavelength higher is the amounts of energy present as one Einstein of light and light at longer the wavelength contains less energy per quantum. Quantum is the measurable unit of light energy. Hence blue light has more energy than red light
400 nm = 71.5 K.cals/mole
500 nm = 57.2 K.cals/mole
600 nm = 47.667 K.cals/mole
EXCITATION AND FLUORESCENCE
In photochemical reactions one photon of a particular wavelength is absorbed by one molecule at a time.
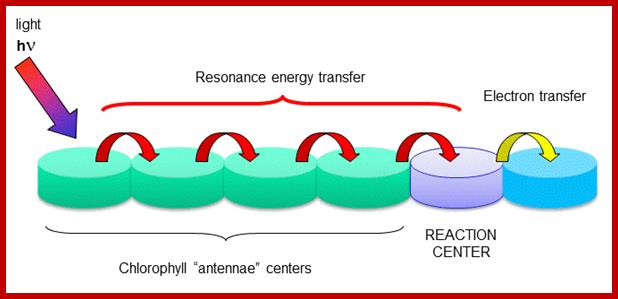
Antenna proteins; The rest of the chlorophyll molecules act as antennas which transfer energy to the reaction centers. Chlorophyll antenna centers; www.kegg.org employees.csbsju.edu
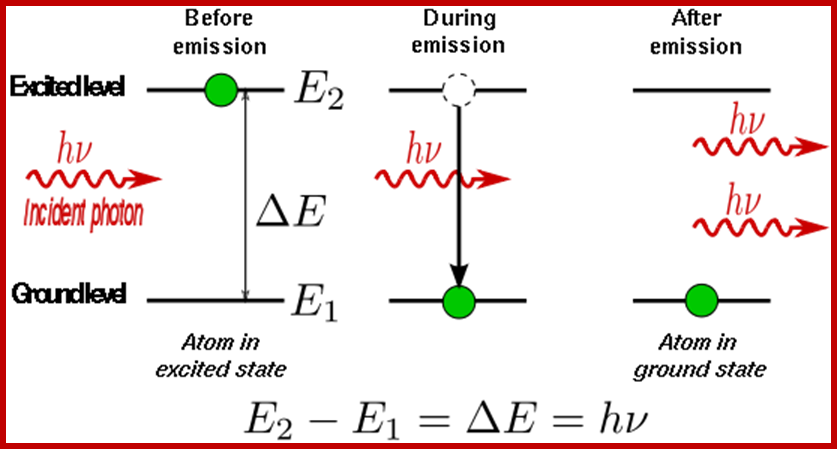
The Pauli exclusion principle forces some electrons to be farther from the nucleus than others, which is why all the electrons in an atom do not simply occupy the 1s orbital. When electrons absorb energy either from light (photons) or from heat, they move farther away from the atomic nuclei but they are only allowed to absorb energy that will land them into specific energy levels. This leads to emission linesand absorption lines.
When an electron is excited, it will not stay that way forever. On average there is a half-life for any particular energy level after which half of the electrons initially in that state will have decayed into a lower state. When such a decay occurs, the energy difference between the level the electron was at and the new level must be released either as a photon or a phonon. When an electron decays without external influence it is said to be due to "spontaneous emission." The phase associated with the photon that is emitted is random. If a number of electrons were put into an excited state somehow and then left to relax, the resulting radiation would be very spectrally limited (only one wavelength of light would be present) but the individual photons would not be in phase with one another. This is also called fluorescence. http://www.thefullwiki.org/
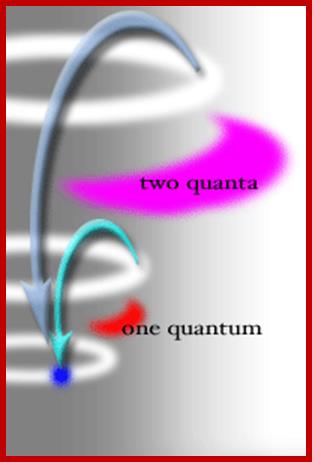
Thus, an excited electron has no option but to give off either 1 quantum or 2 quanta of energy, it cannot give up 1.5 quanta, or 2.3 quanta. Also, the electron can only move to very limited orbitals within the atom; it must end up in an orbital where the wavelength is now uses is "in phase" with itself. These two restrictions limit the quality of the quanta of energy being released by the electron, and thus the nature of the photon of light that rushes away from the LED. http://www.brooklyn.cuny.edu/
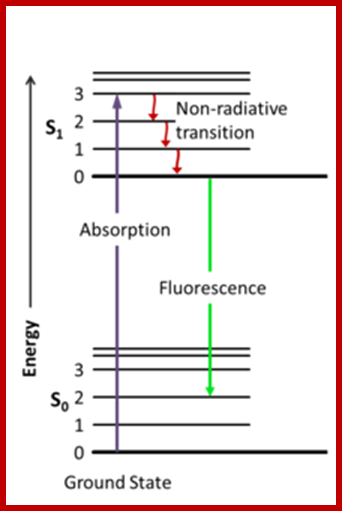
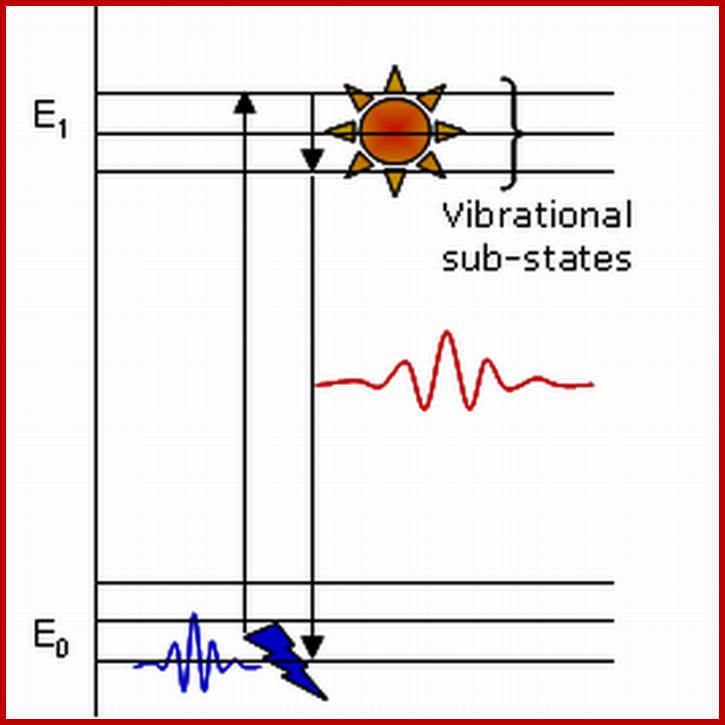
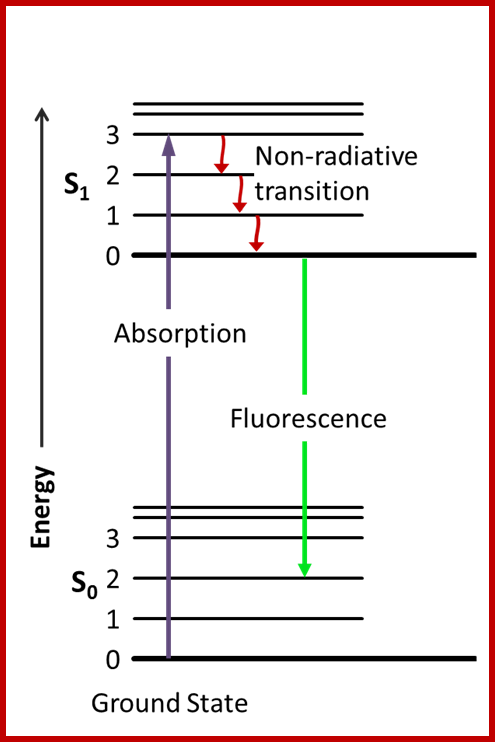
Jablonski diagram; After an electron absorbs a high energy photon the system is excited electronically and vibrationally. The system relaxes vibrationally, and eventually fluoresces at a longer wavelength. Jablonski Diagram; ori-nuxeo.univ-lille1.fr
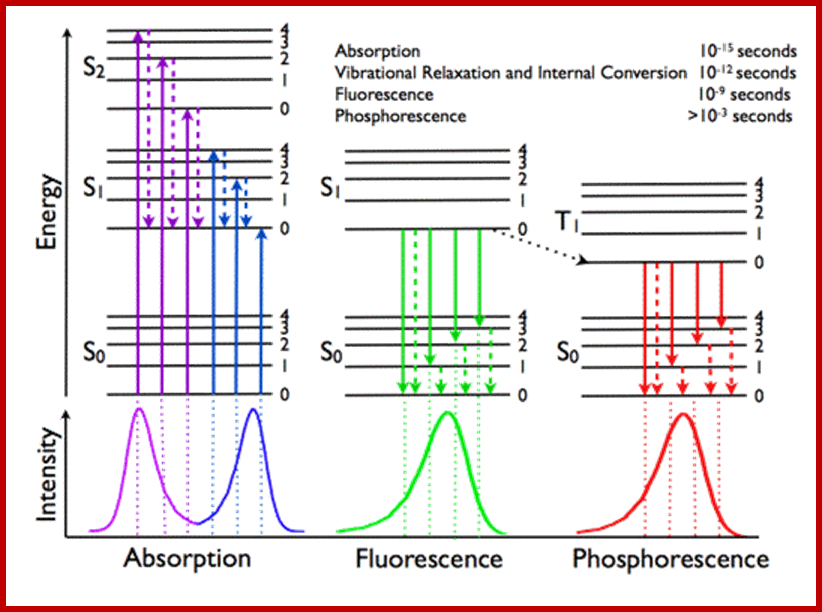
Jablonski diagram representing energy levels and spectra. Solid arrows indicate radiative transitions as occurring by absorption (violet, blue) or emission (green for fluorescence; red for phosphorescence) of a photon. Dashed arrows represent non-radiative transitions (violet, blue, green, red). Internal conversion is a non-radiative transition, which occurs when a vibrational state of a higher electronic state is coupled to a vibrational state of a lower electronic state. In the notation of, for example, S1,0, the first subscript refers to the electronic state (first excited) and the second one to the vibrational sublevel (v = 0). In the diagram the following internal conversions are indicated: S2,4→S1,0, S2,2→S1,0, S2,0→S1,0 and S1,0→S0,0. The dotted arrow from S1,0→T1,0 is a non-radiative transition called intersystem crossing, because it is a transition between states of different spin multiplicity. Below the diagram sketches of absorption-, fluorescence- and phosphorescence spectra are shown; http://www.photobiology.info/
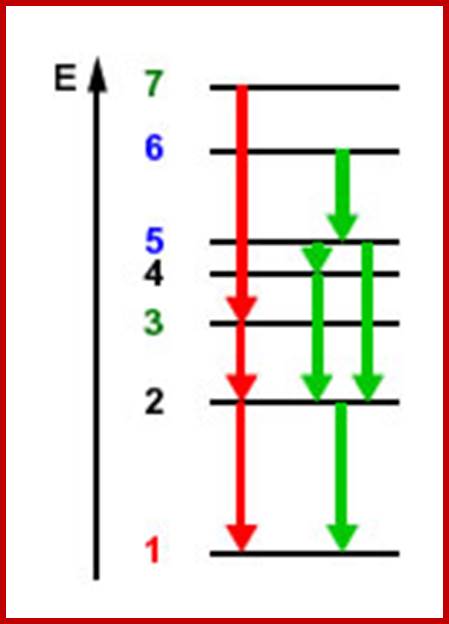
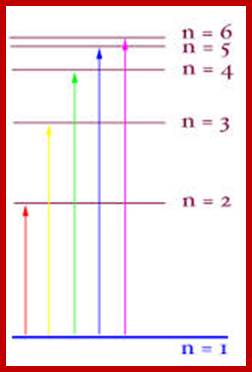
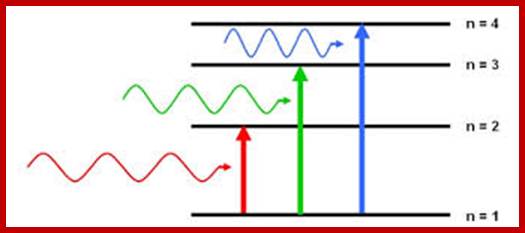
Electrons boosted to excited state, where they remain in the shown there for a fraction of second and fall to the lower orbital with the loss of energy-ultimately to the ground state from where they are boosted.
Molecular orbital states of electrons; www.chemistry.stackexchange.com
Fluorescence; Jablonski diagram; After an electron absorbs a high energy photon the system is excited electronically and vibrationally. The system relaxes vibrationally, and eventually fluoresces at a longer wavelength. en.wikipedia.org
When a photon is absorbed by one multiatomic molecule like chlorophyll the energy level of this molecule is raised to higher state; the photonic energy moves randomly within the molecule and at a particular site, an electron found in the outer orbit, accepts the energy, with its higher energy, it jumps to the next higher orbit. Now the electron said to be in a higher state of energy called excited state or singlet state. Generally, when two electrons are present as pairs in the outer orbit they exhibit opposite spins. According to Pauli’s exclusion principle a pair of electrons in an orbit mentioned above, exhibit magnetic momentum as zero because of the opposite spin. If one of the two electrons is raised to higher state of energy it retains the same spin and such a state is called singlet state.
The same electron can be further raised to the next higher orbital provided the energy is sufficient. The electron in the single state cannot remain not more than 10^-9 sec. It always tends to fall back to its normal orbit, called Ground state. While falling back, electrons change their spin other way. So the spin of the energized electron and the other electron found ground state is rendered same. But according to Pauli’s exclusive principle two electrons with the same spin is forbidden to be present in the same orbit. Thus the energized electron remains suspended for a period of time till it changes its spin into opposite direction.
Such a state of electron is called triplet state and the duration of such metastable state is 10^-6 sec. This time period is quite long enough for a photochemical reaction to occur. If nothing happens in this period, the electron in triplet state changes its spin by internal conversion and falls back to the ground state. The energy thus released is lost as phosphorescent energy or radiant energy.
Now it is known that most of the photochemical reactions take place at the triplet state. This is actually the principle of photochemical act where electrons are boosted to higher energy state, and then they are made to fall back in a stepwise fashion. It is during the descent, the light energy that is released in quantum is used up in the formation of energy rich chemical bonds. Furthermore, it is now clear that in photosynthetic units, electrons are actually transported one by one (unpaired) as in the case of semiconductors. Such a transport is possible only through proteins and its associated chlorophyll molecules. Such events have been detected by observing electron spin resonance signals in isolated chloroplasts even at a temperature as low as O 0C, where enzymatic reactions are totally ruled out.
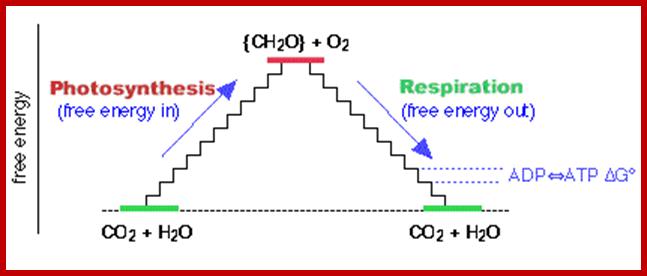
MECHANISM OF PHOTOSYNTHESIS
Photosynthesis is a series of processes of which some photochemical events are independent of temperature and some are temperature dependent enzymatic processes. The structural organization of chloroplasts is so designed the granal structures perform light induced photochemical reactions and then the products are released into the stromatic fluid, where carbon dioxide is fixed into carbohydrates. So, photosynthetic events have been broadly divided into two important sets of reactions called light reactions and dark reactions.
LIGHT REACTIONS
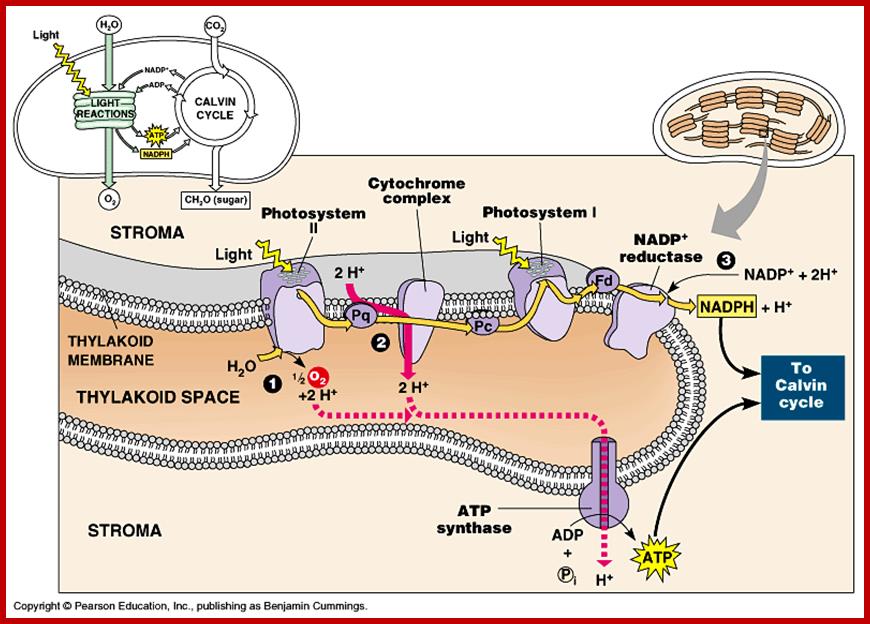
This process is also known as Hill’s Reaction. It takes place mainly in granal and intergranal membranes. Light is primarily responsible for initiating a series of events, in which some are temperature dependent photochemical reactions. In this process three important products are produced i.e. liberation of oxygen, production of reducing power, and ATP synthesis by photophosphorylation.
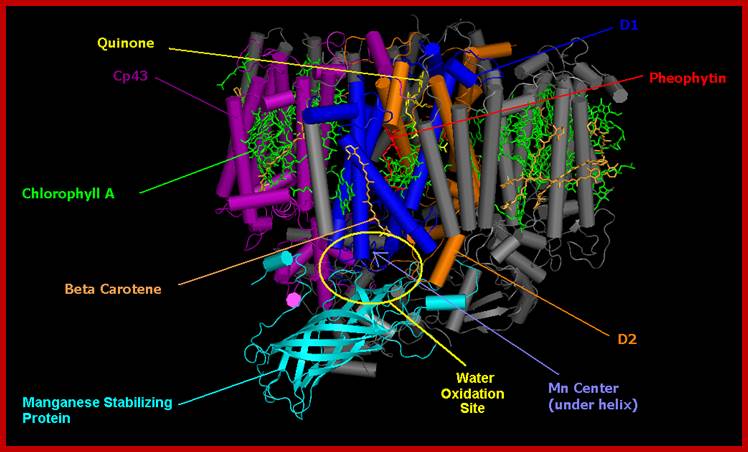
Cyanobacterial photosystem II, Monomer, PDB 2AXT.
Reaction centre PSII; http://rsta.royalsocietypublishing.org/
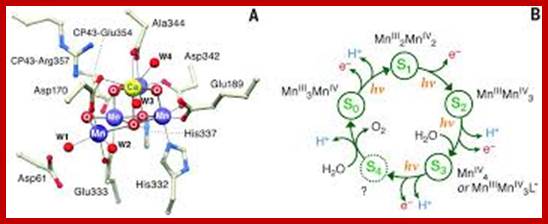
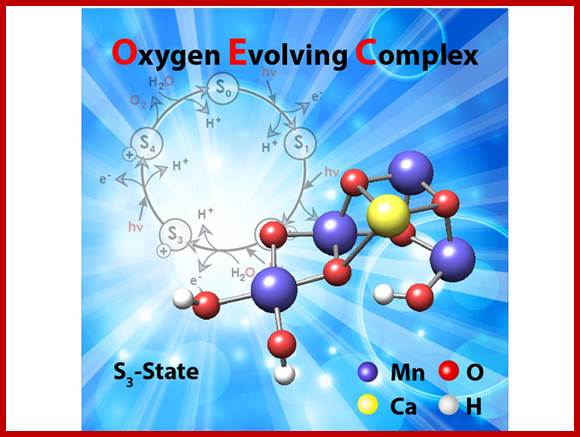
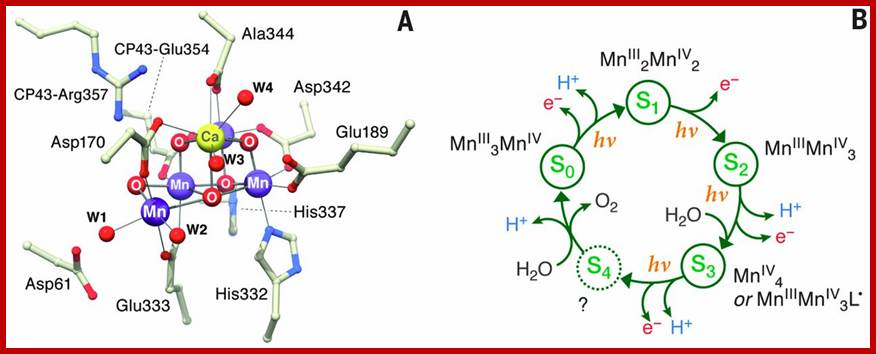
X-ray crystal structure of Oxygen-evolving complex in PhotosystemII prior to O-O bond formation, aMn4O5Ca followed by five’S” states of reaction cycle and in each meta stable state; The water oxidation cofactor of PSII. www.sciencemag.org
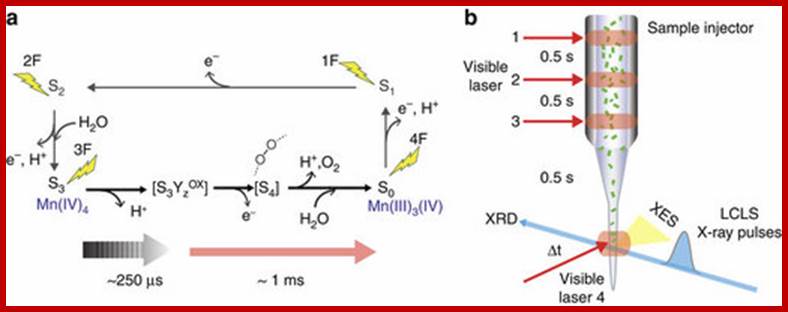
The dioxygen we breathe is formed by light-induced oxidation of water in photosystem II. O2formation takes place at a catalytic manganese cluster within milliseconds after the photosystem II reaction centre is excited by three single-turnover flashes. Here we present combined X-ray emission spectra and diffraction data of 2-flash (2F) and 3-flash (3F) photosystem II samples, and of a transient 3F’ state (250 μs after the third flash), collected under functional conditions using an X-ray free electron laser. The spectra show that the initial O–O bond formation, coupled to Mn reduction, does not yet occur within 250 μs after the third flash. Diffraction data of all states studied exhibit an anomalous scattering signal from Mn but show no significant structural changes at the present resolution of 4.5 Å. This study represents the initial frames in a molecular movie of the structural changes during the catalytic reaction in photosystem II. Jan Kern, et al, www.nature.com
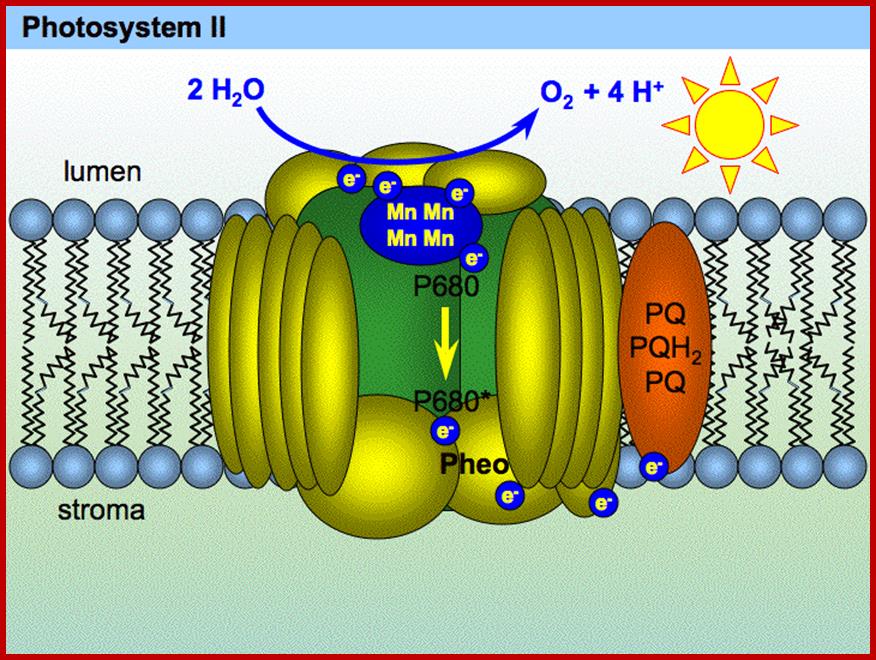
LIBERATION OF OXYGEN:
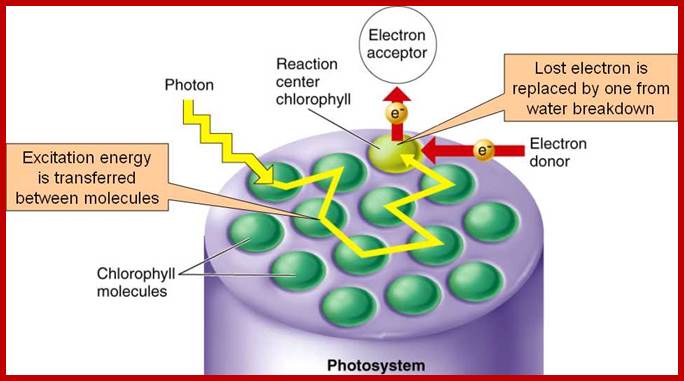
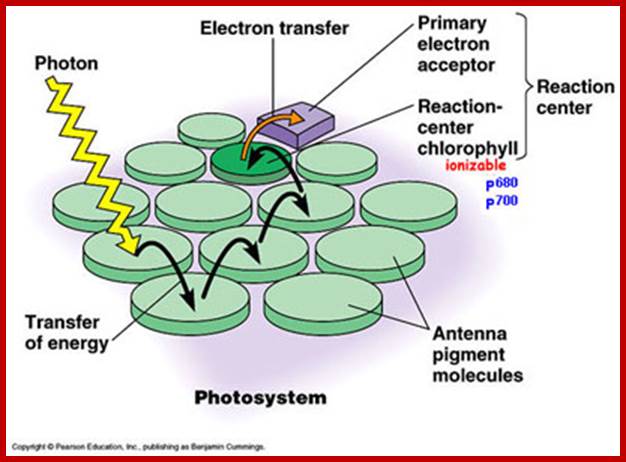
As the solar radiations strike the leaf surface, the chlorophyll molecules with their antenna protein complexes present in Quantosome absorb the photons belonging to specific wavelengths and transfer the same from molecule to molecule by a mechanism called resonance. Thus chlorophyll molecules with the absorbed light energy are raised to higher state of energy called excited state.
The excited chlorophyll protein complexes as they are surrounded by water molecules exert electrical effect on water. So the water molecules are cleaved into the respective ions like hydroxyl ions (OH) and hydrogen ions (H+). This process is called photo ionization.
The hydroxyl ions thus produced are immediately accepted by Z-protein Mn2+ complex found in PS II system. The OH ions are transitorily accepted by Mn2+ ions. Each Mn2+ can take up two OH ions, so totally four OH are now bound two energized Z-Mn proteins via arginine residues. Then OH ions by rearranging their orbitals react to produce a molecule of H2O and two atomic oxygen. As atomic oxygens are highly unstable they immediately combine to produce molecular oxygen. In these rearrangement reactions of four hydroxyl ions four electrons are also released.
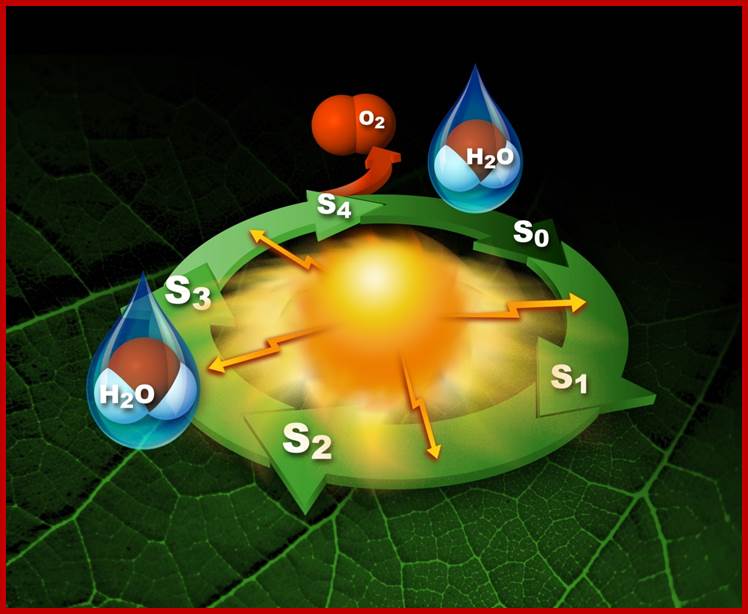
Photosytem II utilizes a water-splitting manganese-calcium enzyme that when energized by sunlight catalyzes a four photon-step cycle of oxidation states (S0-to-S3). When S3 absorbs a photon, molecular oxygen (O2) is released and S0 is generated. S4 is a transient state on the way to S0. (Image courtesy of SLAC National Accelerator Laboratory) - See more at: http://newscenter.lbl.gov/2014/07/09/femtosecond-snapshots-of-photosynthetic-water-oxidation/#sthash.QzJeWit4.dpuf; http://newscenter.lbl.gov/
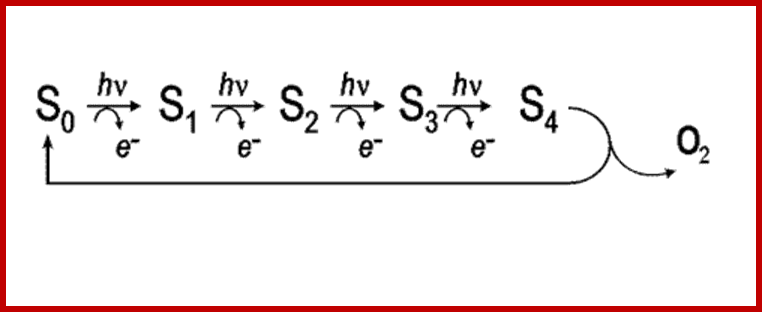
The structure of the manganese cluster as it is found in nature and prior to O-O bond formation. In the background, the water-splitting cycle with the intermediate states S0 to S4. Credit: MPI for Chemical Energy Conversion; http://phys.org/
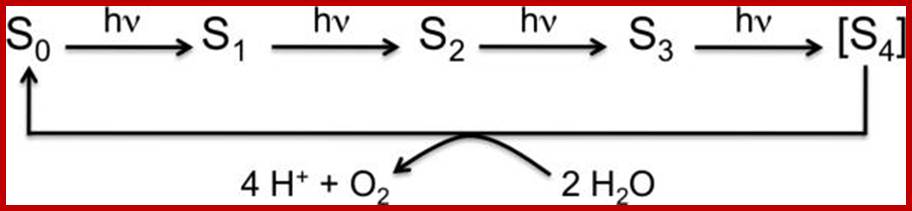
Reaction cycle of water oxidation at the Mn4CaO5 cluster in PS II. http://www.ncbi.nlm.nih.gov/
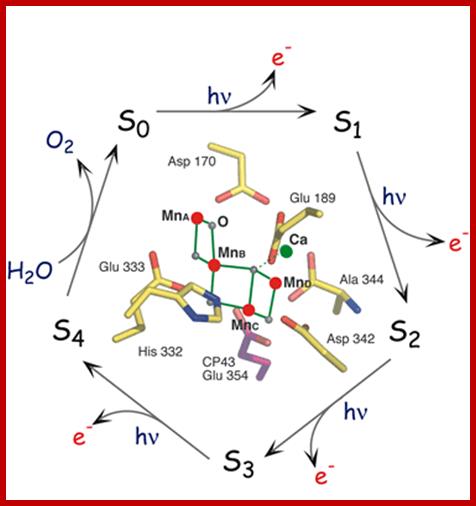
This image portrays the water-splitting catalytic cycle with the Mn4Ca structure in the middle; www2.lbl.gov
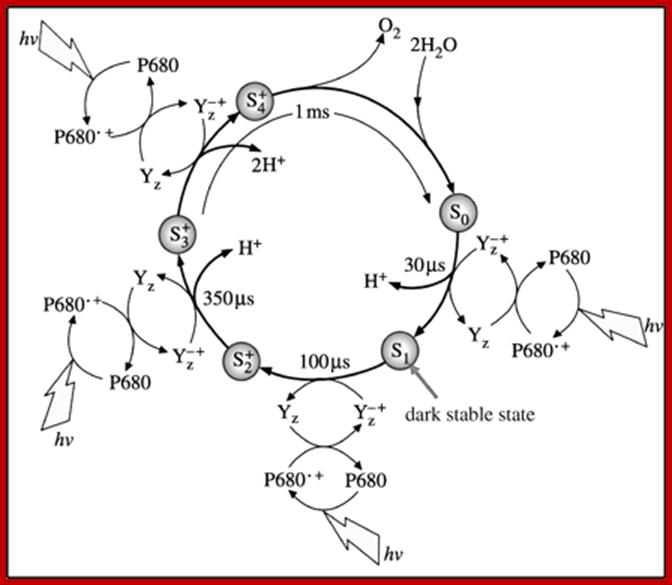
The S-state cycle showing how the absorption of four photons of light (hν) by P680 drives the splitting of two water molecules and formation of O2 through a consecutive series of five intermediates (S0, S1, S2, S3and S4). Protons (H+) are released during this cycle except for the S1 to S2 transition. Electron donation from the Mn3Ca2+O4 cluster to P680+ is aided by the redox active tyrosine TyrZ, abbreviated to YZ here. Also shown are half-times for the various steps of the cycle. http://rsta.royalsocietypublishing.org/
Interestingly, in this reaction four OH ions are accepted at four equital steps by Z-Mn complex. Each step requires a photon. Hence the Z-Mn2+ Complex goes through different states called So, S1, S2, S3. Such reactions have been confirmed by detecting the electron spin resonance (ESR) signals.
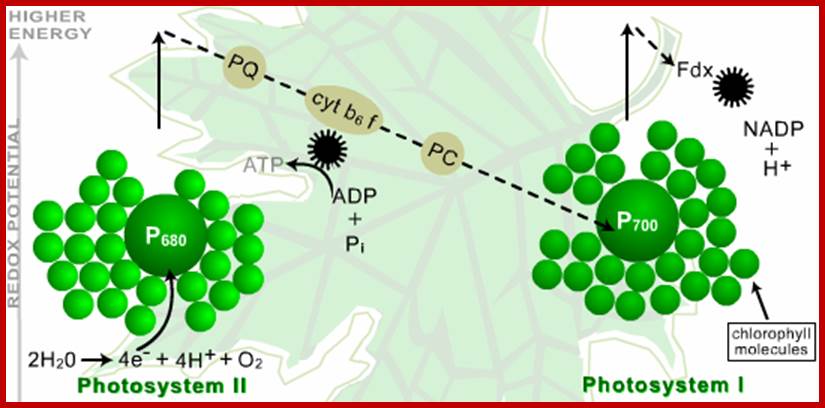
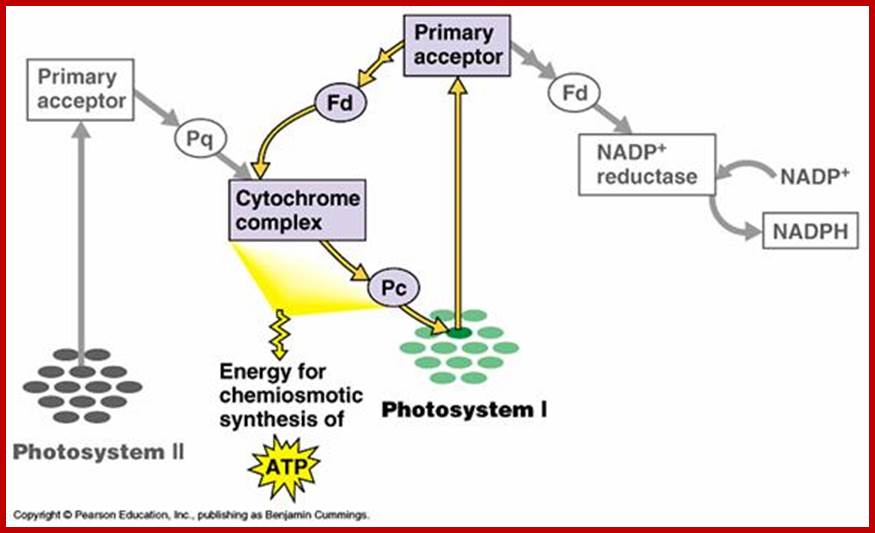
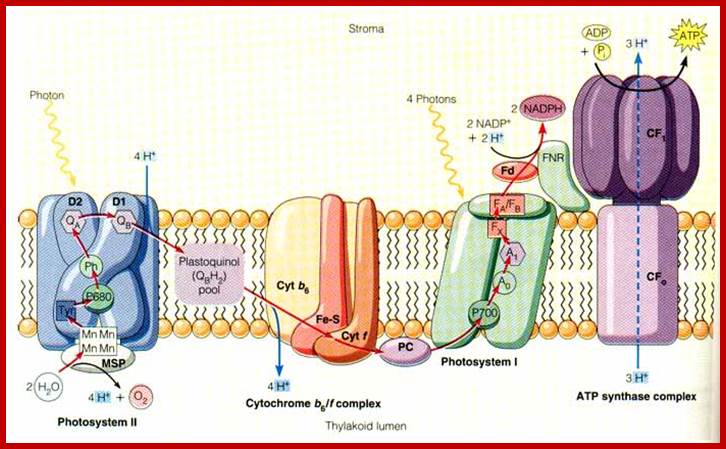
GENERATION OF REDUCING POWER
Reduction of NADP to NADPH+H takes place at the outer surface of thylakoids and the NADPH+H+ thus produced is called reducing power. In thylakoids after absorbing light by PSI electrons are transferred from molecule to molecule and ultimately they reach the reaction centre i.e. Chl.a 700. There are two such chla.700 molecules for every photosynthetic unit. They are associated with their respective protein complexes. With the excitons reaching Chl.a 700, the pigment gets excited, because at C-O site (in all probabilities) an electron is raised to higher orbital where they may undergo internal conversion and state triplet state for a period of 107 to 108sec.
www.photobiology.info
Photosystem I typically includes 13 polypeptide chains, more than 60 chlorophyll molecules, a quinone (vitamin K1), and three 4Fe-4S clusters. The total molecular mass is more than 800 kd. Photosystem II is only slightly less complex with at least 10 polypeptide chains, more than 30 chlorophyll molecules, a nonheme iron ion, and four manganese ions. The absorption of a second photon and the movement of a second electron down the path from the special pair completes the two-electron reduction of QB from Q to QH2. http://plantphys.info/
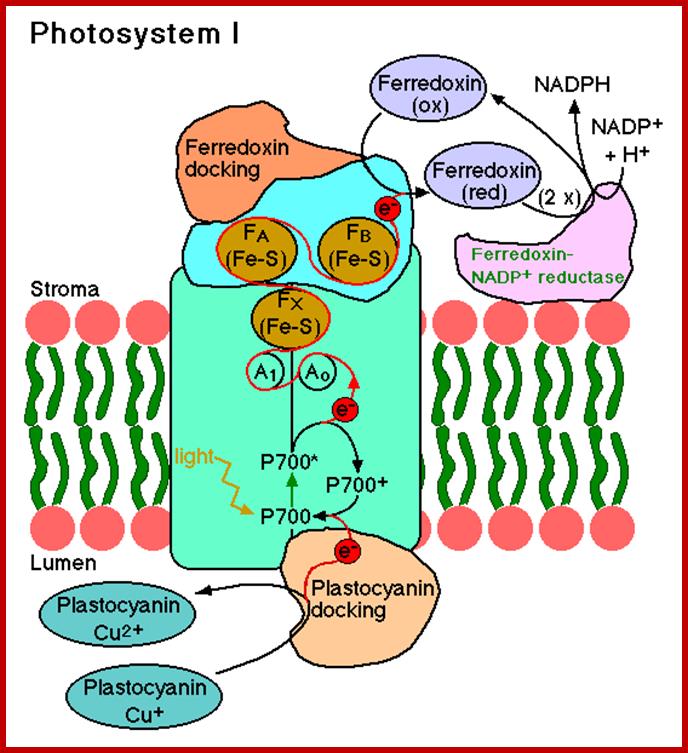
It is at this juncture, the high energy electrons are accepted by ferrodoxin reducing substance. From two Chl. 700 molecules two electrons are transferred to FRS and the same are transferred to non-heme iron containing protein called ferrodoxin. Then these electrons are transferred to NADP through NADP reductase. Simultaneously, the energy rich NADP accepts two protons (H+). These reactions results in the formation of reducing power called NADPH+H. Though some amount of energy is lost during the transfer of electrons from Chl A 700 to NADP, a significant amount of solar energy is conserved in NADPH+H as chemical energy.
Most importantly, in this process, as the electrons are lost from the Chl. 700 molecules they are rendered electrically positive. In this state Chl. 700 molecules cannot remain for a long time. But the photosynthetic units i.e. PS I and PS II are organized in such a way the positively charged Chl. 700 molecules receive electrons from Chl.A 680 from PS II system through an electron transport chain. So Chl.700 molecules are rendered neutral till they go through another set of reactions leading to the formation of another Molecule.
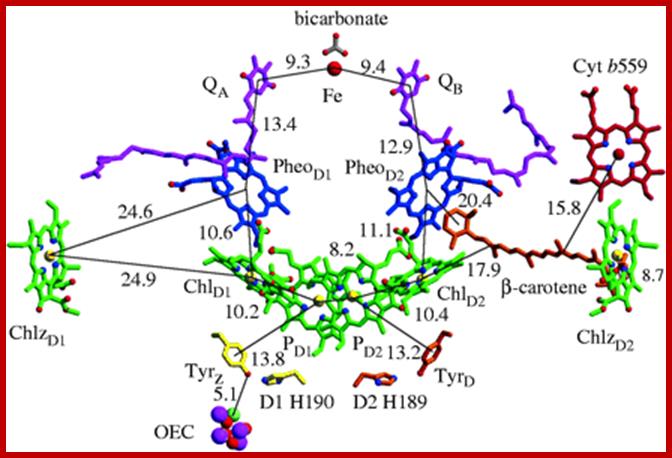
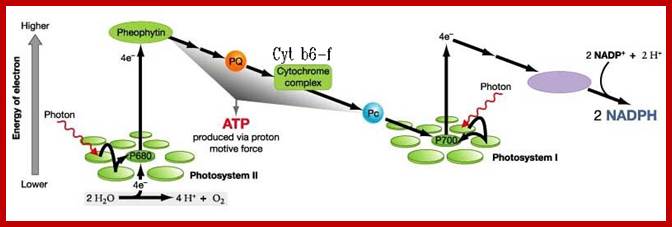
The PS II system also absorbs light energy and the same is conveyed to Chl.A 680 active centers where the electrons are boosted to a higher state of energy then the energy rich electrons are accepted by pheophytin. Pheaophytin is similar to chlorophylls but lack Mg in the center of porphyrin ring.
From pheophytin electrons are transported down the hill through an electron transport chain towards Chl.A 700. In this process, Chl. 680 molecules are rendered positively charged. However, the Chl. 680 molecules receive electrons from Z-Mn2 (OH) protein mediated oxygen liberation reactions. Thus Chl.a 680 are rendered neutral. It should be noted that the flow the electrons from Chl. A 700 to NADP and from Chl.a 680 to Chl.a 700 plus from 2nMn2+ complex to Chl.a 680 is non cyclic process.
PHOTOPHOSPHORYLATION AND ATP SYNTHESIS
Energy trapping and conversion and conservation of the same in the form of chemical energy is one of the fascinating aspects of photosynthesis. Besides, producing energy rich NADPH molecules, Quantosomes, i.e. PS I and PS II are also capable of generating high energy chemical bonds in ATP, while transporting electrons from one acceptor to the other. Such a process is called photophosphorylation. In fact, plants have evolved to produce energy rich molecules like ATP by two mechanisms namely non cyclic photophosphorylation and cyclic photophosphorylation.
NON CYCLIC PHOTOPHOSPHORYLATION
Non cyclic photophosphorylation takes place in thylakoids where both PS I and PS II are in close proximity with each other and function co-ordinatingly, but now it is revealed that in granal stacks most of the photosystems found are PSII.
The flow of electrons in this case is discontinuous because the electrons boosted to higher state of energy from Chl. A 700 of PS I which are found in stromal lamellae, are accepted by FRS and then they are transferred to NADP via FD. The electrons lost from dimer Chl. A 700 are replaced by electrons boosted by Chl. 680 molecules of PS II ?.
The high energy electrons from Chl. 680 dimers are first accepted by pheophytin complex, and then they are transferred from one acceptor to another sequentially finally to Chl.a 700.
Co-factor for oxidation of water PSII (© 2014 American Association for the Advancement of Science All Rights http://ibitecs.cea.fr/Reserved.).
It is while the electrons flow from Chl.a 680 to Chl.a 700, because of difference in the redox potential between Cyt. B. 559 and Cyt.C, a significant amount of electronic energy is liberated and the same is used up by ATP synthetase to bring about the synthesis of an ATP molecule. For every pair electron flow from one molecule of ATP can be synthesized in this process. However, recent investigations suggest that there is one more site for non – cyclic photophosphorylation between Z-Mn2+ complex and Chl. A 680.
CYCLIC PHOTOPHOSPHORYLATION

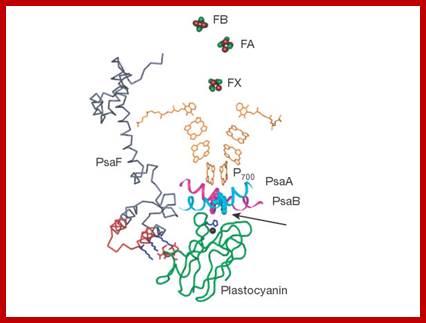
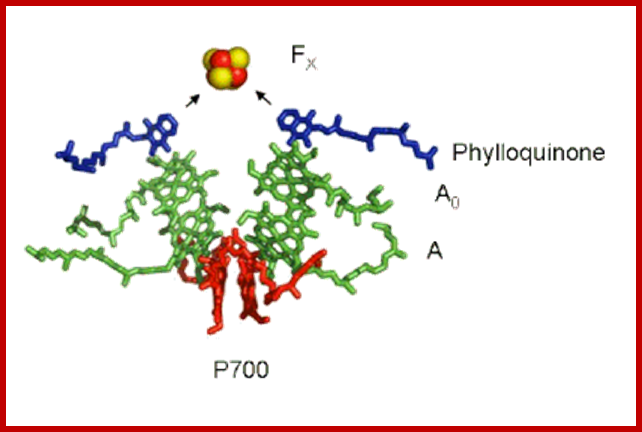
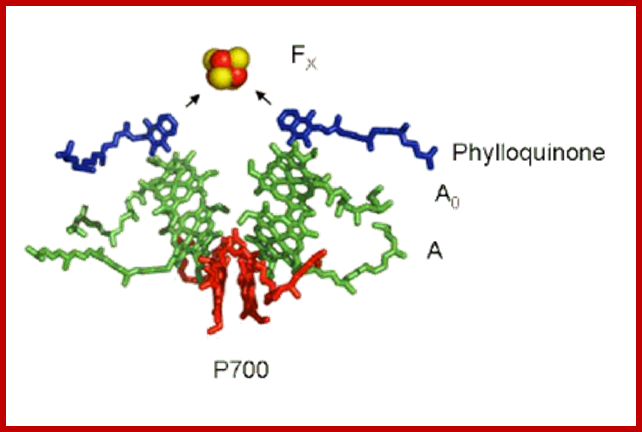
Cofactors of the PSI Reaction Center: The "special pair" of chlorophyll-a molecule called P700 is shown in red. The other chlorophyll a species that are part of the pathway of electron transfer are shown in green, and the phylloquinone molecules are shown in dark blue. The primary electron acceptor is the four iron four sulfur cluster called FX, shown here as a red (iron) and yellow (sulfur) cube. http://photobiology.info/
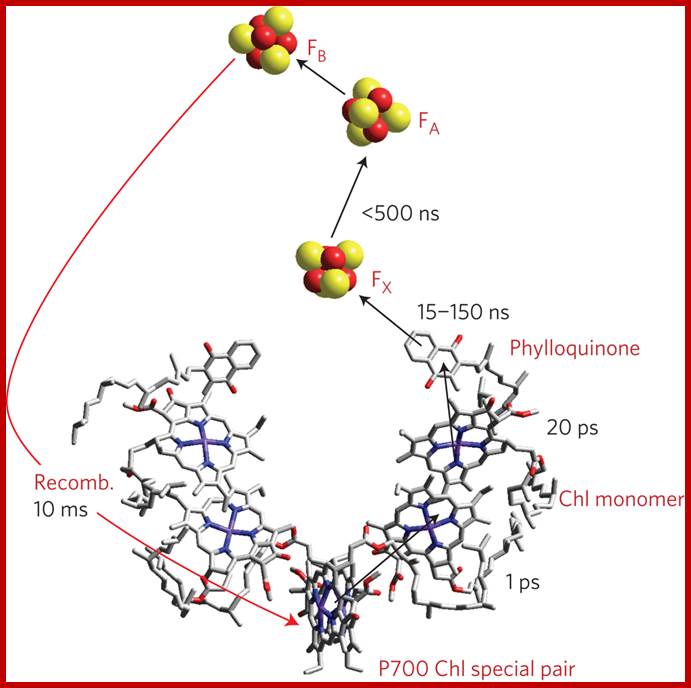
http://www.nature.com/ Reaction centre PS I
After photoexcitation of an electron in the chlorophyll (P700) special pair, the electron is transferred to three [4Fe–4S] iron–Sulphur clusters. Photosystem II (PSII) is the water splitting enzyme of photosynthesis. Its appearance during evolution dramatically changed the chemical composition of our planet and set in motion an unprecedented explosion in biological activity. Powered by sunlight, PSII supplies biology with the ‘hydrogen’ needed to convert carbon dioxide into organic molecules. The questions now are can we continue to exploit this photosynthetic process through increased use of biomass as an energy source and, more importantly, can we address the energy/CO2 problem by developing new photochemical technologies which mimic the natural system? (Critical review, 82 references).
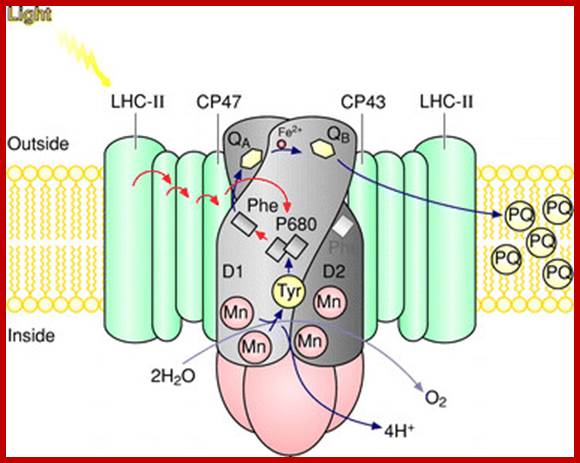
Schematic drawing of the photosystem II antenna system and reaction centre in the thylakoid membrane. D1 and D2 are the core proteins of photosystem II reaction centre. Photosystem II uses light energy to remove electrons from water, resulting in the release of oxygen and protons. The electrons from water are transferred via redox cofactors in the protein complex to form reduced plastoquinone. (Mn)4, manganese cluster involved in removing electrons from water; P680, reaction centre chlorophyll of photosystem II; Pheo, pheophytin; QA, a bound plastoquinone; QB, plastoquinone that binds and unbinds from photosystem II; Tyr, a tyrosine residue in photosystem II (Yz); PQ, a pool of mobile plastoquinone molecules. The antenna complexes are: LHC-II, peripheral light-harvesting complex of photosystem II; CP47 and CP43, chlorophyll–protein complexes of 47 and 43 kDa, respectively. https://www.researchgate.net
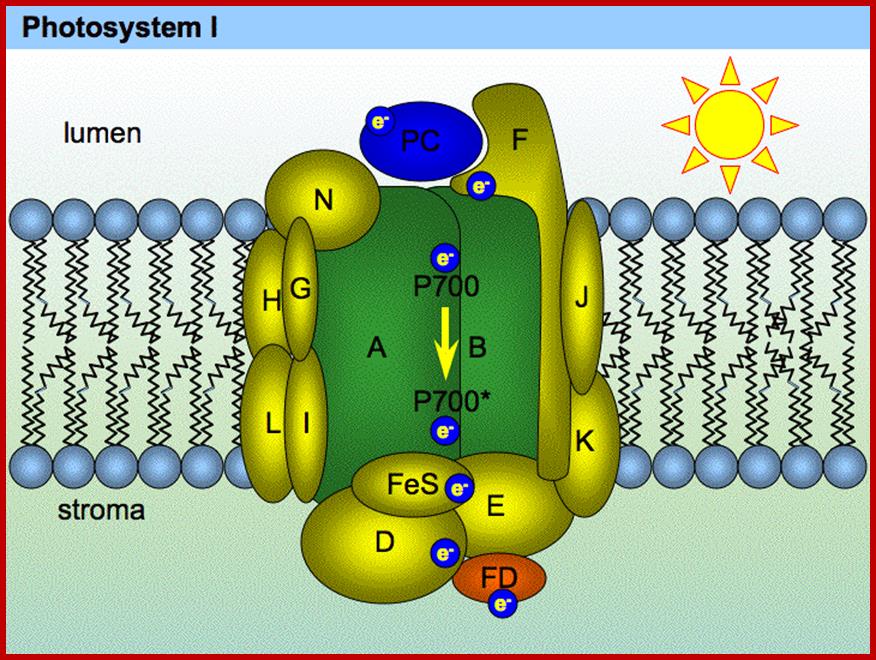
Photosystem I (PSI) [2] is an integral membrane protein complex that useslight energy to mediate electron transfer from plastocyanin to ferredoxin. Linear electron transport in PSI produces both ATP and NADPH, while cyclic electron transport drives the production of ATP but not the production NADPH.[3] The PS I system comprises more than 110 co-factors, significantly more than photosystem II.[4] These various components have a wide range of functions. The electron transfer components of the reaction center of PSI are a primary electron donor P-700 (chlorophyll dimer) and five electron acceptors: A0 (chlorophyll), A1 (a phylloquinone) and three 4Fe-4S iron-Sulphur centers: Fx, Fa, and Fb.[5]
Two main subunits of PS I, PsaA and PsaB, are closely related proteins involved in the binding of P700, A0, A1, and Fx. PsaA and PsaB are both integral of 730 to 750 amino acids that seem to contain 11 transmembrane segments. The Fx 4Fe-4S iron-Sulphur center is bound by four cysteines; two of these cysteines are provided by the PsaA proteinand the two others by PsaB. The two cysteines in both proteins are proximal and located in a loop between the ninth and tenth transmembranesegments. A leucine zipper motif seems to be present [6] downstream of the cysteines and could contribute to dimerization of psaA/psaB. The terminal electron acceptors, FA and FB, are located in a 9 kDa protein called PsaC.[ https://en.wikipedia.org
The antenna complex is composed of molecules of chlorophyll and carotenoids mounted on two proteins.[11] These pigment molecules transmit the resonance energy from photons when they become photo excited. Antenna molecules can absorb all wavelengths of light within the visible spectrum. The number of these pigment molecules varies from organism to organism. For instance, the cyanobacterium Synechococcus elongatus (Thermosynechococcus elongatus) has about 100 chlorophylls and 20 carotenoids, whereas spinach chloroplasts have around 200 chlorophylls and 50 carotenoids.[4][12] Located within the antenna complex of PS I are molecules of chlorophyll called P700 reaction centers. The energy passed around by antenna molecules is directed to the reaction center. There may be as many as 120 or as few as 25 chlorophyll molecules per P700
The P700 reaction center is composed of modified chlorophyll a that best absorbs light at a wavelength of 700nm, with higher wavelengths causing bleaching.[14] P700 receives energy from antenna molecules and uses the energy from each photon to raise an electron to a higher energy level. These electrons are moved in pairs in an reduction process from P700 to electron acceptors. P700 has an electric potential of about -1.2 volts. The reaction center is made of two chlorophyll molecules and is therefore referred to as a dimer.[11] The dimer is thought to be composed of one chlorophyll a molecule and one chlorophyll a' molecule (p700, Webber). However, if P700 forms a complex with other antenna molecules, it can no longer be a dimer. Two main subunits of PS I, PsaA and PsaB, are closely related proteins involved in the binding of P700, A0, A1, and Fx. PsaA and PsaB are both integral membrane proteins of 730 to 750 amino acids that seem to contain 11 transmembrane segments.
www.Photobiology .Info
www.majordifferences.com; www.uic.edu
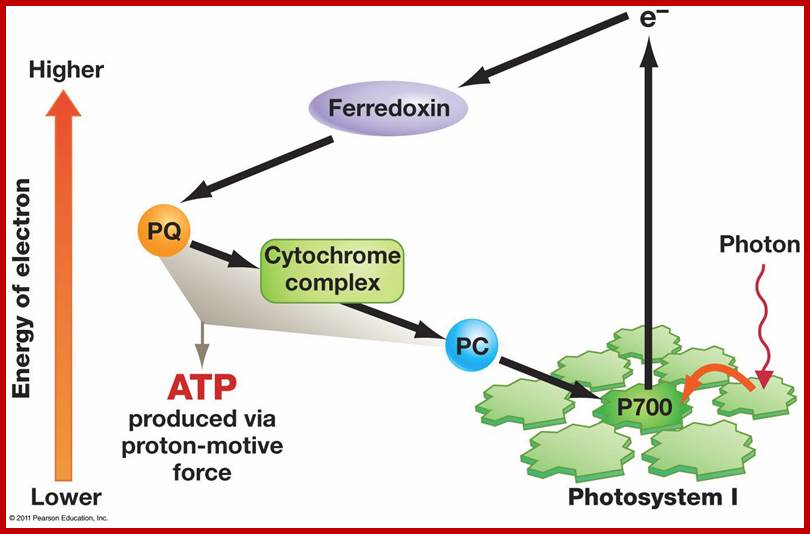
This process occurs in the intergranal lamellae or stromal lamellae which contain mostly PS I systems and its associated electron transporting component. When the solar energy is absorbed by antenna Chlorophyll protein complexes found in PS I system, the energy is used to boost electrons from Chl.a 700 dimers. The energy rich electrons are immediately accepted by FRS, whose red ox potential is 0.6 Ev. From FRS the energy rich electrons (in pails) are transferred to FD and then to cyt. B6 is quite substantial and the energy released in this step is used to produce a molecule of ATP. From cyt.B6 electrons are transferred to cyt.f and from cyt.f to Chl.a 700 through plastocyanin. Again the energy that is released between cyt.b6 and Cyt.f redox couple is used in the formation of another molecule of ATP. The rest of the energy that is not utilized in the ATP synthesis is lost as thermal energy. In this scheme electrons complete one cycle, that is why this process is called cyclic photophosphorylation (Fig. 11.18 & 10)
MECHANISM OF ATP SYNTHESIS
The release of energy during electron transport in itself is into enough for the formation of a terminal bond between ADP and Pi. The formation of ATP requires the activity of ATP synthetase. This enzyme consists of protein components called CF 1 and HFI, the former is located on the outer surface of the thylakoid and intergranal lamellae and the later is buried is the core of the membrane. Many theories have been proposed to explain the synthesis of ATP, of which Peter Mitchell’s chemiosmotic hypothesis and Boyer’s conformational chemical coupling hypothesis are very significant.
ATP synthase at the right side of the top figure is an exception ‘molecular machine’ for ATP production; http://www.uncommondescent.com/
Chemiosmosis mode of ATP synthesis; www.myrome.org
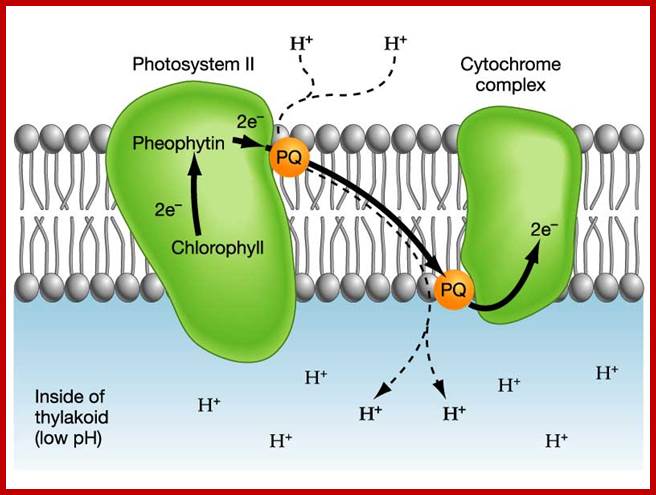
According to chemiosmotic hypothesis, the hydrogen ions produced during photoionization (it need not be) are accepted up by plastoquinones which also accepts electrons donated by pheophytin. The reduced plastoquinones i.e. PQH2 shuttles within the membrane and discharges only H+ into the space found in the thylakoid sac, but electrons on the other hand are channeled towards Chl.a 700 through electron transport chain. Because of charge separation and accumulation of H+ ions, the liquid present in the thylakoid space, becomes more acidic and the liquid present at the outer surface of the thylakoid more basic.
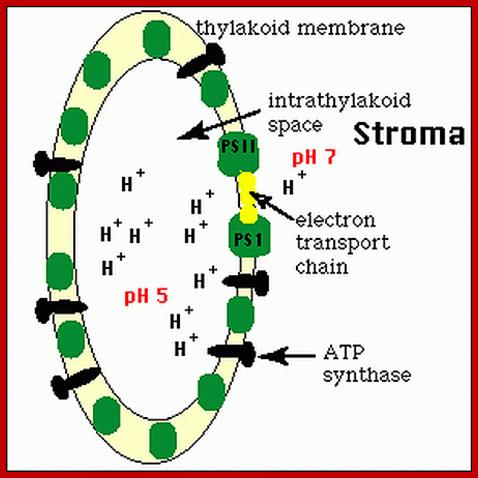
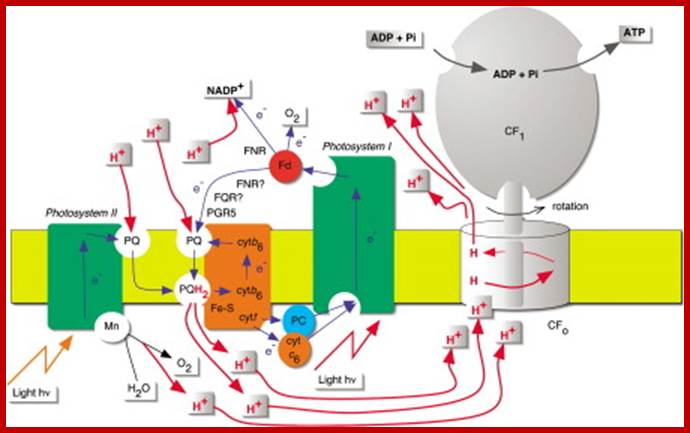
Light-driven electron transport is coupled to ATP synthesis in chloroplasts. While the nature of the coupling and the structures of key components are now known, there has long been disagreement over pathways of electron transport. Recent results now put an old idea back on the agenda—cyclic electron transport around photosystem-I http://www.sciencedirect.com/
Photosynthesis–From Light to ATP; Electron transport (e−) (blue) is arranged vectorially in the chloroplast thylakoid membrane (yellow). Proton (H+) (red) translocation from the chloroplasts stroma (above the membrane) into the lumen (below the membrane) establishes a proton motive force that couples electron transport to ATP synthesis. The implied stoichiometry 3H+/e− is for noncyclic electron transport alone (cf. Figure 2). FQR is a hypothetical ferredoxin-Quinone-oxidoreductase. Junge's animations Rotary ATP Synthase and From Light to ATP are recommended viewing,
Figure - Coupling Quanta, Electrons, Protons, and ATP Combined cyclic and noncyclic photophosphorylation, assuming H+/3ATP = 14. Stoichiometries are depicted for four electrons transferred from H2O to NADP+ (cf. Figure 1). This gives one O2 molecule and two NADPH molecules. Three ATP molecules will be made, provided photosystem I recycles one electron in order to contribute two protons to the proton motive force; http://www.biologie.uni-osnabrueck.de/Biophysik/Junge/overheads.html
ATP synthase; Molecular Machine to Generate ATPs:
ATP synthase works at the expense of ‘transmembrane electrochemical proton potential difference’ to generate ATPs and it in different form acts as ATPase; It holds good for all bacteria (Prokaryotes) and Animals and Plants (Eukaryotes)
ADP + Pi + Energy -˃ ATP.
ATP -˃ ADP + Pi + 7.3 K. cal /mole.
Depending upon the organisms the structurally exhibit minor variations, but their function is same. The spherical structure that sits on membrane is called F1 and the base that is buried in the membrane is called Fo. The number of protein subunits vary from 20 -22.
Based on organisms and structural features they are classified into F-, A-, P-, and E- ATPases-
F-type ATPase: "F-type ATPase" or FoF1 ATPase (bacterial) is just another name for ATP synthase; letter "F" comes from "phosphorylation Factor". F-ATPases are present in bacteria, mitochondria and chloroplasts. Their major function in most cases is ATP synthesis at the expense of the transmembrane electrochemical proton potential difference. In vitro F-type ATPases can operate in both directions depending on conditions. Bacterial ATP synthase or FoF1-Atpase a multisubunit complex of 9x20nm (dimater X length) dimension and ~530kDa mol.wt is located in the bacterial PM, is a molecular dynamo. The Fo is tight ring of 10-14 subunits called C. And the gamma subunit the rotor is not held by C ring
- A-type ATPases: A-type ATPases were found in Archaea, their function is similar to that of F-type ATP synthase, but structurally they are very similar to V-type ATPases.
- V-type H+-ATPases; V-type H+-ATPases were initially found in eukaryotic Vacuoles. Their primary function is ATP-driven proton (or Na+) pumping to acidify the vacuol interior.
- P-type ATPases (sometimes named E1-E2 ATPases); P-type ATPases pump a variety of ions across the membrane and are found in bacteria and in many eukaryotic cell organelles.
- E-type ATPases (do not mix with E1-E2 ATPases!); E-type ATPases is a family of enzymes that hydrolyze Extracellular ATP (see Herbert Zimmermann's Ecto-ATPase webpage for details)
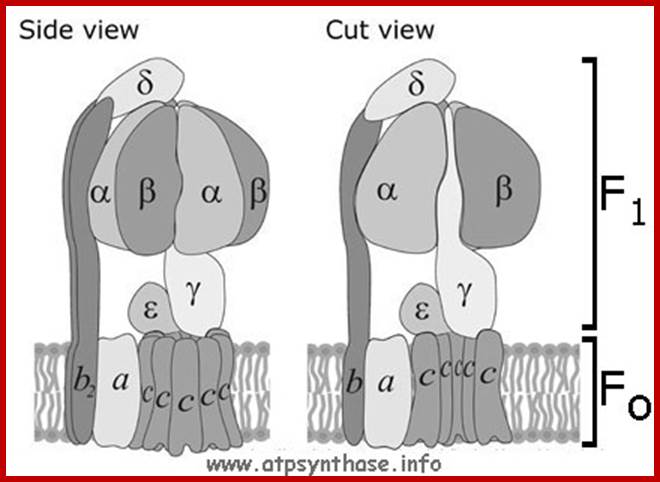
The catalytic portion of ATP synthase (F1) is formed by Alpha 3 Beta 3 hexamer with Gamma subunit inside it and Epsilon attached to the Gamma. Subunit Delta is bound to the "top" of the hexamer and to subunits b. The hydrophobic transmembrane segment of subunit b is in contact with subunit a. Subunits Gamma and Epsilon of the catalytic domain are bound to the ring-shaped oligomer of c-subunits. Proton translocation take place at the interface of subunits a and c. www.atpsynthase.info
Fo-Driven by the proton motive force, protons are transferred through the FO portion of the enzyme. This transfer drives the rotation of the c-subunit oligomer ring relative to the a and b subunits.
- The rotation is passed to Gamma and Epsilon subunits that are bound to the c-subunit oligomer ring. The rotation of asymmetric Gamma subunit mechanically causes conformational changes in Alpha 3 Beta 3 -hexamer. Each 120 degrees of the Gamma subunit rotation forces one of 3 catalytic sites located at Alpha-Beta interface into an opened conformation. Freshly synthesized ATP molecule is released, and phosphate and ADP are bound instead. High affinity of the opened site to phosphate impairs rebinding of ATP and favors ADP binding.
- Rotation goes further, Gamma subunit turns another 120 degrees forcing the next site into the opened conformation, and the ADP and phosphate bound to the previous opened site are occluded and ATP synthesis takes place. The ATP molecule formed is released when the Gamma subunit makes one 360 degrees turn and once again opens the site. www.atpsynthase.info
How many catalytic sites does the enzyme have?
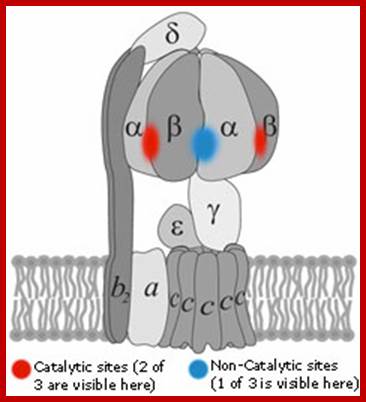
The answer is three. Not five. However, the total number of the nucleotide-binding sites is six, three of them being non-catalytic. Each site is located on the interface between subunits Alpha and Beta. Larger part of each catalytic site is composed from amino acid residues of the respective Beta-subunit, while each non-catalytic site is situated mostly on the respective Alpha subunit. The role of the non-catalytic sites is probably regulatory, they are not necessary for the catalysis. Occupation of the non-catalytic sites by nucleotides was shown to increase the enzyme activity. It is also possible that binding of nucleotides to the non-catalytic sites facilitate the enzyme assembly in the cell.
There is strong evidence that in bacteria of Bacillus genera the Epsilon subunit also can bind one nucleotide. So in Bacillus ATP synthase there are 7 nucleotide binding sites! webkit-text-stroke-width: 0px;"> www.atpsynthase.info
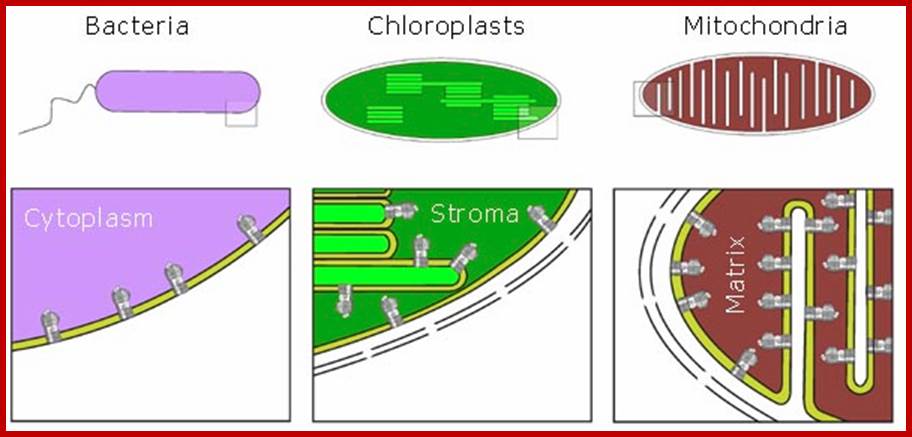
Position of ATP synthases are shown in the diagrams of bacteria, plastids and mitochondria. web kit-text-stroke-width;". www.atpsynthase.info
Bacterial ATP synthase or FoF1-Atpase a multisubunit complex of 9x20nm (dimater X length) dimension and ~530kDa mol.wt is located in the bacterial PM, is a molecular dynamo. The Fo is tight ring of 10-14 subunits called C. And the gamma subunit the rotor is not held by C ring
This creates proton motive force and a charge in the membrane potential. In order to neutralize these charges, the protons are pumped out by the hydrogen transporting protein located at the base of the ATP synthetase complex. It is during the transfer of protons the energy generated is used in the formation of a chemical bond between ADP and Pi. With the release of OH into the thylakoid space and H into the stromatic fluid the charges are neutralized. In fact, Jogendorf provided a very good evidence for chemiosmotic mode of ATP synthesis. When isolated chloroplasts are first suspended in an acidic pH medium and then if they are transferred to a medium with basic pH, containing ADP and Pi, energy rich ATP molecules are generated.
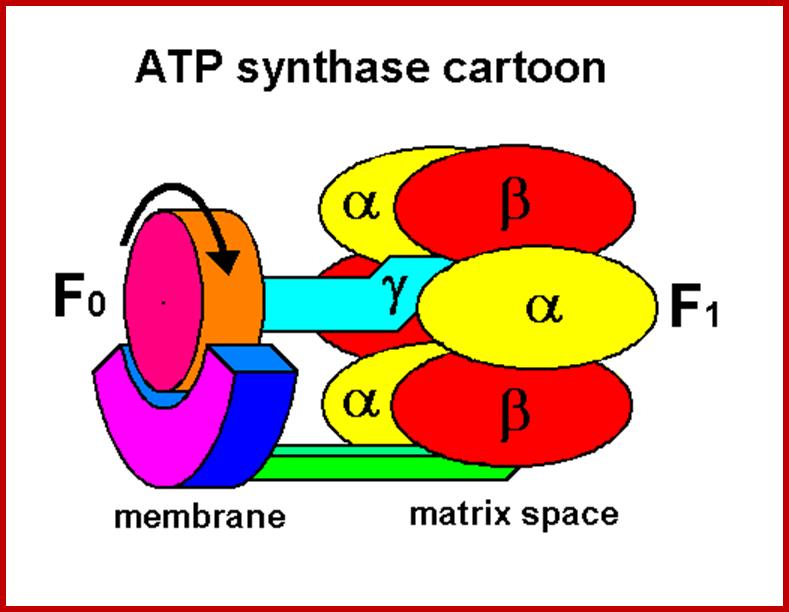
http://www.bmb.leeds.ac.uk/illingworth/6form/
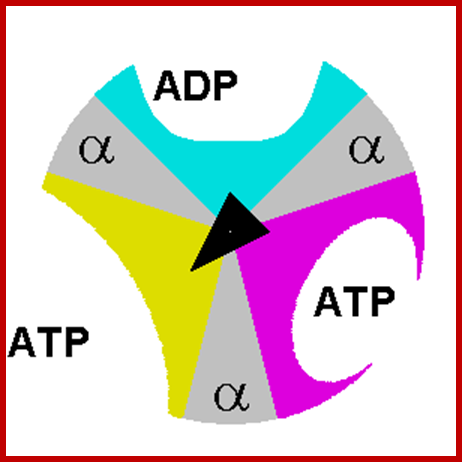
http://www.bmb.leeds.ac.uk/illingworth/6form/
In recent years, chemiosmotic hypothesis has been questioned because intergranal lamellae containing PS I system also synthesize ATP by cyclic photophosphorylation. This process takes place in spite of the intergranal lamellae do not contain inner space as found in thylakoid membranous sacs. Furthermore, the time required for generating such strong proton motive force requires at least few milliseconds, but in actuality the synthesis of ATP takes place in microseconds. In recent years new microelectrodes have been developed which could measure slightest change in pH in shortest possible time. Such studies suggested that chemiosmotic hypothesis is no more tenable but it is too early to deny the hypothesis once and for all. However to day every scientist has agreed to chemiosmotic hypothesis in generating ATP molecules not only in chloroplasts but also in mitochondria.
In recent years, chemiosmotic hypothesis has been questioned because intergranal lamellae containing PS I system also synthesize ATP by cyclic photophosphorylation. This process takes place in spite of the intergranal lamellae do not contain inner space as found in thylakoid membranous sacs. Furthermore, the time required for generating such strong proton motive force requires at least few milliseconds, but in actuality the synthesis of ATP takes place in microseconds. In recent years new microelectrodes have been developed which could measure slightest change in pH in shortest possible time. Such studies suggested that chemiosmotic hypothesis is no more tenable but it is too early to deny the hypothesis once and for all. However, to day every scientist has agreed to chemiosmotic hypothesis in generating ATP molecules not only in chloroplasts but also in mitochondria.
* Creation & the ATP SYNTHASE Motor
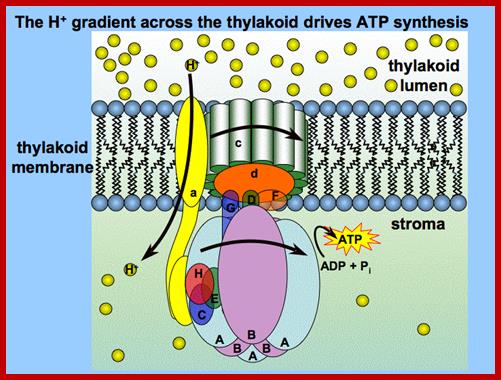
www.plantphys.info
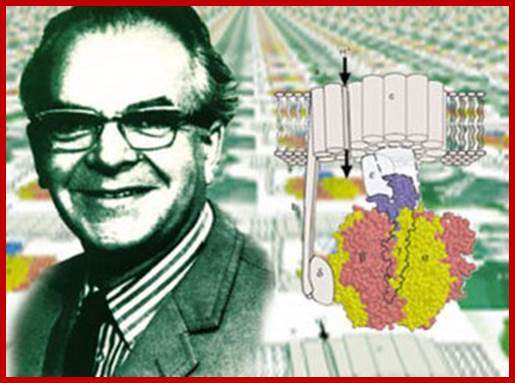
Peter Mitchell and ATP synthase enzyme@1999 nature publishing group orgel, L; Are you serious Mitchell? Nature402,17 (1999), All rights reserved. The proton gradients that power respiration are as universal as the genetic code itself, giving an insight into the origin of life and the singular origin of complexity. Why are cells powered by proton gradients; Nick Lane, Ph.D. http://www.nature.com/
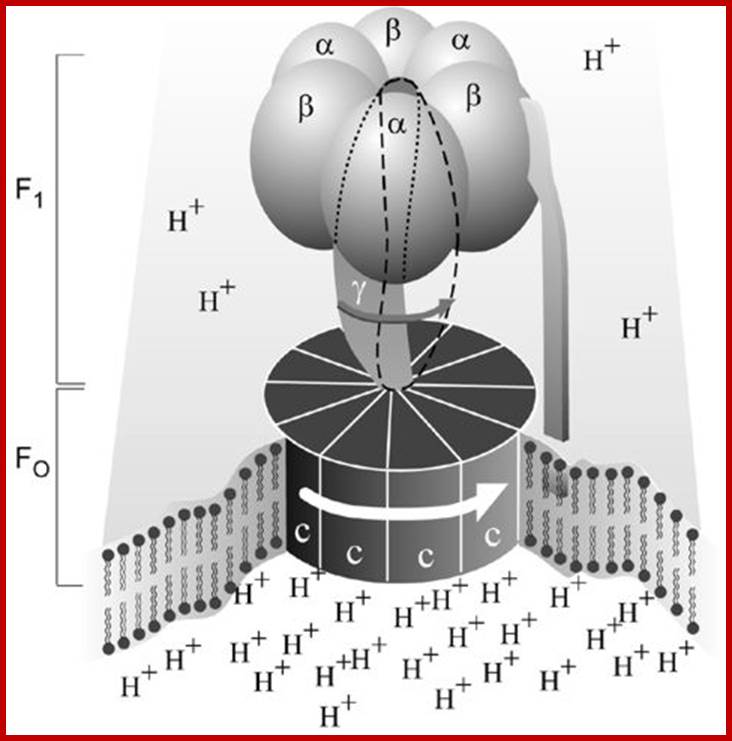
Prof. Paul D. Boyer,Dr. John E. Walker and Prof.Jens C.Skou; www.nobelprize.org
Simplified picture of ATP synthase
The Fo part through which hydrogen ions (H+ ) stream is located in the membrane. The F1 part which synthesizes ATP is outside the membrane. When the hydrogen ions flow through the membrane via the disc of c subunits in the Fo part, the disc is forced to twist around. The gamma subunit in the F1 part is attached to the disc and therefore rotates with it. The three alpha and three beta subunits in the F1 part cannot rotate, however. They are locked in a fixed position by the b subunit. This in turn is anchored in the membrane. Thus the gamma subunit rotates inside the cylinder formed by the six alpha and beta subunits. Since the gamma subunit is asymmetrical it compels the beta subunits to undergo structural changes. This leads to the beta subunits binding ATP and ADP with differing strengths
But Boyers conformational chemical coupling hypothesis appears to be more valid. For his proposal many evidences have come up. He was also awarded a Nobel Prize, much later than Peter Mitchell received Nobel Prize. Accordingly, during the electron transport along the electron transport chain energy released causes conformational change in the contractile proteins found in the granal membrane. The presences of such contractile proteins have been identified in chloroplast membranes and they are believed to be associated with ATP synthetase. The consisted contractile protein later relaxes by releasing the energy to ATP synthetase which by virtue of its activity, traps this energy, as a result the enzyme itself undergoes conformational change where it brings the enzyme bound ADP and inorganic Pi close to each other to bring about an anhydride bond formation between ADP and Pi to produce ATP molecules. This process takes place in a few microseconds and added to this, it does not require any kind of proton motive force for the synthesis of ATPs. Boyer’s hypothesis is now more or less accepted by most of the workers in the field of Bioenergetics. Yet chemiosmotic process of loading thylakoid space with hydrogen ions is a fact.
DARK REACTION OR CARBON PATHWAY
Dark reaction, which is also called by names like carbon pathway, Blackman’s reaction, C3 pathway etc, is a temperature dependent enzymatic process, where the products of light reactions like ATP and NADPH are used in fixing carbon dioxide into simple carbohydrates like glucose. However, the mechanism of carbon fixation or dark reaction varies in different plants. Accordingly, three different pathways have been recognized so far. They are Calvin’s cycle or C3 pathway. Hatch and Slack pathway C4 pathway and the third Crassulacian acid metabolic (CAM) pathway. Most of the dicot members (with certain exceptions) and majority of monocots operate Calvin’s pathway. But tropical grass members and some dicots like Atriplex, etc. exhibits C4 pathway. The succulents and some orchids on the other hand show CAM pathway as a method of carbon fixation.
CALVIN’S CYCLE OR C3 PATHWAY
Utilizing radioactive tracers like 14C bicarbonate and auto radiographic techniques, Calvin and his associates have elucidated various biochemical steps involved in this pathway. This involves three major steps viz. Carboxylation, Reduction and Regeneration (CRR).
CARBOXYLATION
Biochemical analysis of photosynthetic cells like chlorella and others show that a 5 carbon sugar called Ribulose diphosphate (RUDP or RuBP) acts as an acceptor of carbon dioxide. When the cells are illuminated the concentration of RUBP decreases and Phosphoglycerate (PGA) increases. On the contrary, when light is switched off, the concentration of RUBP increases while the concentration of PGA goes down in the chloroplasts. Based on these experimental evidences RUBP has been identified as the primary acceptor of CO2.
www.biology.arizona.edu; www.algaeenergy.weebly.com
Initially Ribulose monophosphate (RUMP) is converted to RUBP by a kinase enzyme. The five carbon RUBP with binding of enzyme undergoes a transitory stage which facilitates the binding of CO2. The resultant 6C compound, still bound to the enzyme surface, is highly unstable and breaks up into two 3C compounds called phosphoglycerate. They are the first stable products of carbon fixation.

The enzyme that is responsible for this reaction is called RUBP carboxylase, which is known to be the most abundant protein is nature because more than 50% of the leaf proteins is RUBP carboxylase. In chloroplasts this enzyme is located as a granular particle on the outer surface of thylakoid membrane. It consists of 4 smaller subunits coded for by nuclear genome and 4 larger subunits coded for by plasto genome.
Carbon fixation-https://www.studyblue.com
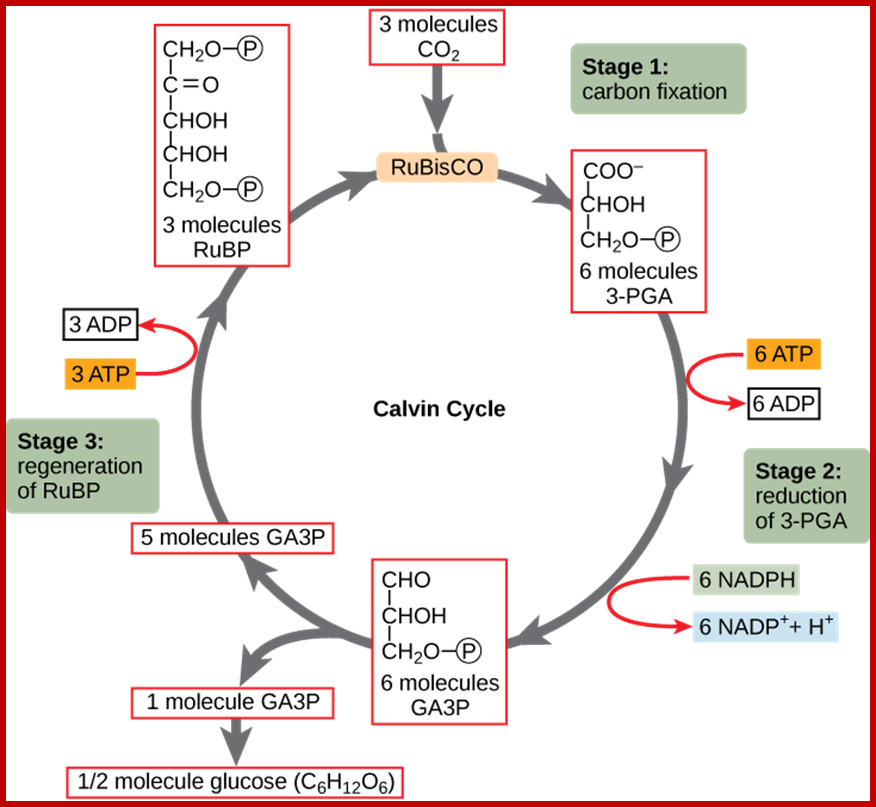
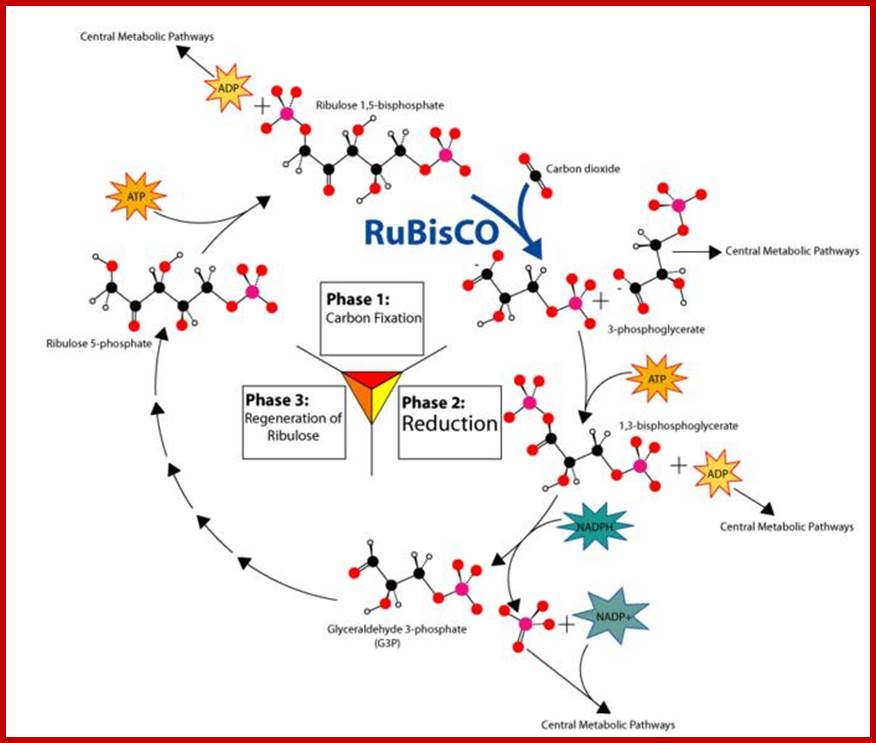
The Calvin cycle has three stages. In stage 1, the enzyme RuBisCO incorporates carbon dioxide into an organic molecule, 3-PGA. In stage 2, the organic molecule is reduced using electrons supplied by NADPH. In stage 3, RuBP, the molecule that starts the cycle, is regenerated so that the cycle can continue. Only one carbon dioxide molecule is incorporated at a time, so the cycle must be completed three times to produce a single three-carbon GA3P molecule, and produce a
Six times to six-carbon glucose molecule; ;www.cnx.org
Source: “The Calvin Cycle.” Boundless Biology. Boundless, 09 Mar. 2016. from https://www.boundless.com/biology/textbooks/boundless-biology-textbook/photosynthesis-8/the-light-independent-reactions-of-photosynthesis-82/the-calvin-cycle-377-11603/ https://www.boundless.com
REDUCTION
Phosphoglycerate is then converted to an energy rich di phosphoglycerate by the activity of an enzyme called phosphoglycerate kinase. One molecule of ATP is used in this process. Then the DIPGA is reduced to phosphoglyceraldehyde by the action of glyceraldehyde phosphate dehydrogenase. NADPH + H that is synthesized during light reaction donate hydrogen to this reaction. In fact, in the above reactions, energy stored in ATP and NADPH+H is transferred to PGALD.
REGENERATION
The process of regeneration involves a series of inter linked reactions in which the initial substrate i.e. RUMP used in Carboxylation step is regenerated; simultaneously glucose is also synthesized as an ultimate product of CO2 fixation. In this process some of the PGALD are converted to dihydroxyacetone phosphate by an enzyme called isomerase.
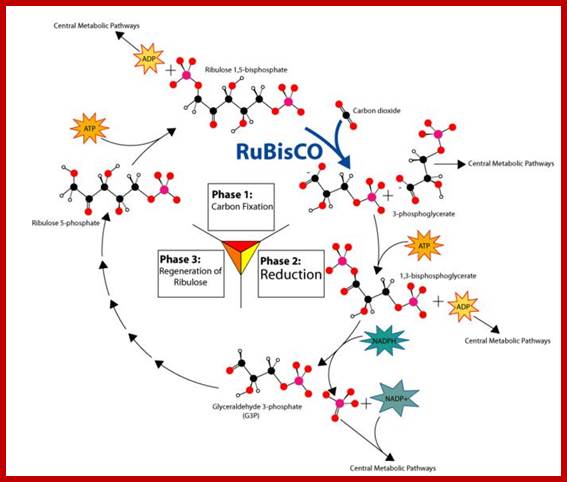
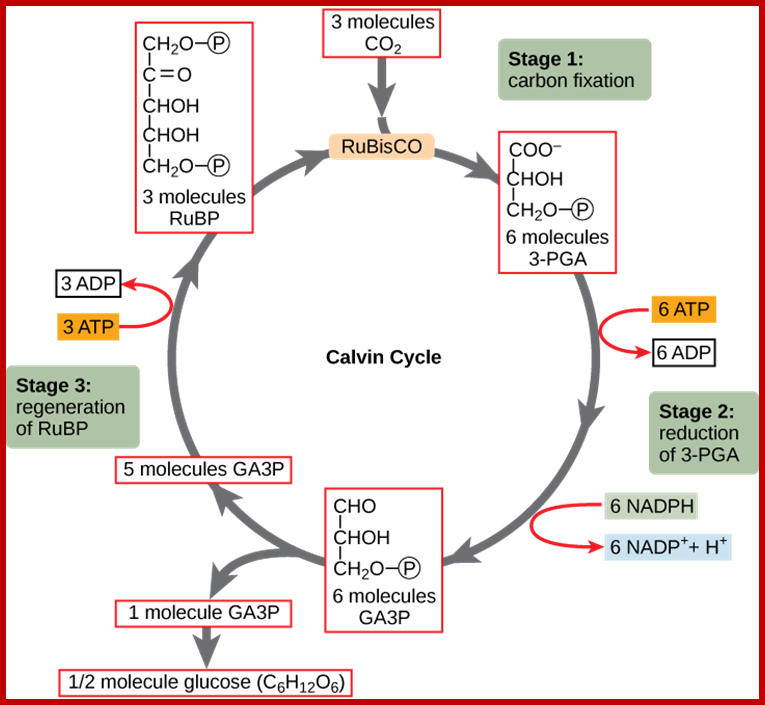
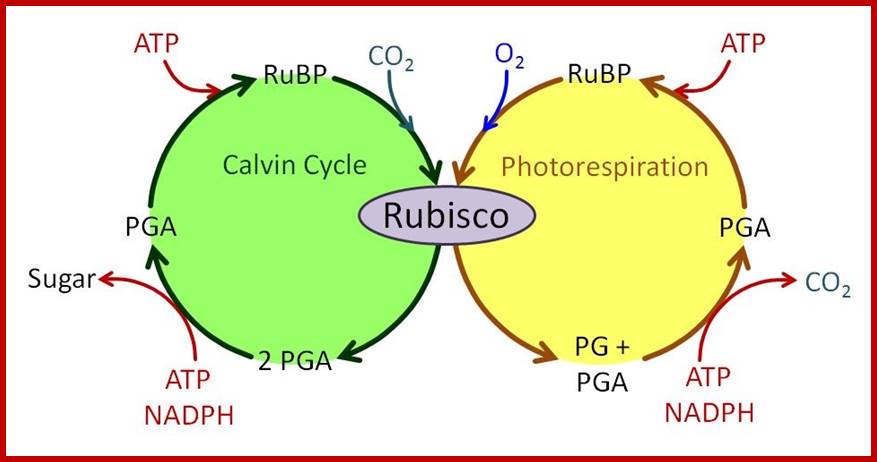
www.cnx.org
Then 3 molecules of PGALD and 3 molecules of DiHAP (DAP) are condensed head-to-head to synthesize 3 molecules, of 6C fructose 1, 6 diphosphate. The enzyme responsible for this reaction is aldolase.
The fructose diphosphate then undergoes dephosphorylation at 1st carbon position by the action of phosphatase enzymes.
Two of the three fructose-6P thus produced combine with two PGALD to generate 2 xylulose 5 P and 2Erythrose 4, P molecules. This reaction is performed by the enzyme called transkelotase.
The 2 Erythrose phosphates fuse with 2 molecules of DHAP to produce 2 molecules of seven carbon Sedoheptulose diphosphate. The enzyme that brings about this reaction is Aldolase. The Sedoheptulose diphosphates are then dephosphorylated at first carbon atom by the phosphatase enzyme.
The transkeletolase enzymes brings about another rearrangement reaction between 2 Sedoheptulose 7(P) and 2 PGALD to generate 2 molecules of ribose 5 (P) and 2 molecules of xylulose 5(P).
The ribose phosphor-isomerase and the xylulose epimerase convert ribose 5, p and xylulose 5P into Ribulose 5P respectively.
One of the Fructose 6P remained in the reaction (3) is converted to glucose 6P by glucophosphate isomerase. The glucose may dephosphorylated by another phosphatase enzyme.
The overall reactions in this regenerative steps results in the formation of 6 molecules of RUMP and one molecule of glucose as the end product carbon fixation.
7PGALD + 5 DHAP –> 6 RUMP + C6H12O6 + 6 Pi
HATCH AND SLACK PATHWAY OR C4 PATHWAY
http://journal.frontiersin.org/Journal/10.3389/fpls.2013.00085/full
Tropical grass members like crab grass, sugar cane, Zea mays, etc., while performing photosynthetic reactions operate light reactions similar to that of other dicot members belonging to C3 class of plants. But with regard to carbon fixation, they fix carbon first into a C4 organic acid, which is later used in the formation of glucose. Nevertheless these plants use both C3 cycle as well as C4 pathway but in different0 cell types found in the same leaf.
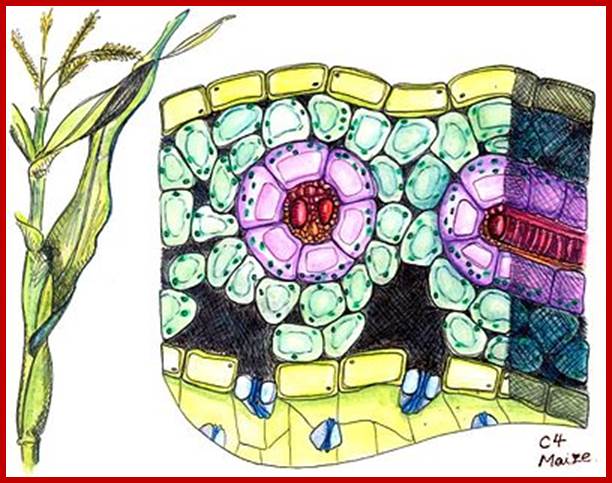
Cross section of a C4 plant, specifically of a maize leaf. Kranz anatomy shown. Drawing based on microscopic images courtesy of Cambridge University Plant Sciences Department.; http://en.wikipedia.org/wiki/C4_carbon_fixation
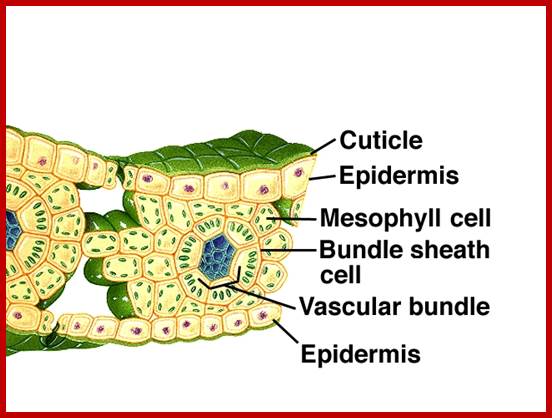
www2.mcdaniel.edu; iws.collin.edu
Grass leaves contain a layer of bundle sheath cells around vascular bundles, thus separates mesophyll cells from the veins. Detailed studies suggest that the chloroplasts of bundle sheath cells differ structurally and functionally from that of chloroplasts found in the mesophyll cells.
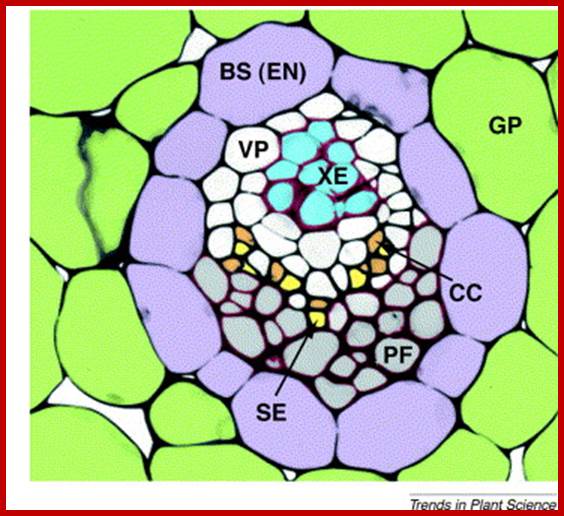
Macromolecular trafficking in the phloem; www.cell.com
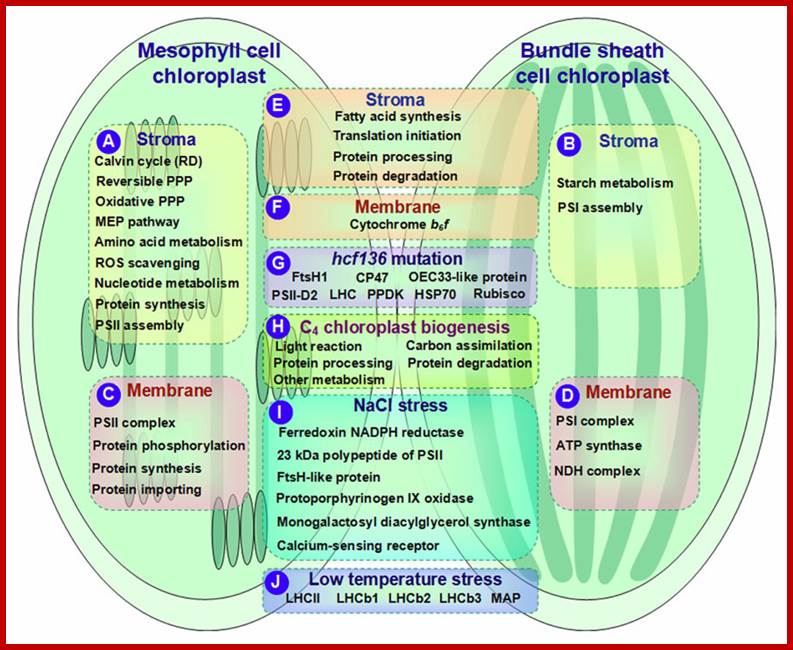
Schematic presentation of cell-specific or stress-responsive pathways and proteins in maize chloroplasts revealed from proteomics studies. (A, B). Preferential metabolic pathways in the stroma of M and BS, respectively; (C, D) Preferential metabolic pathways and protein complexes in the chloroplast membrane of M and BS, respectively; (E,F) Metabolic pathways and protein complexes equally distributed in the chloroplast stroma/membrane of M and BS; (G) Differentially expressed proteins in chloroplasts of maize hcf136 mutant in comparison to wild-type; (H) Dynamics of metabolic pathways during maize chloroplast biogenesis; (I) Salt-responsive proteins in maize chloroplasts; (J) Low temperature-responsive proteins in maize chloroplasts. ATP, adenosine triphosphate; HSP, heat shock protein; LHC, light harvesting complex; MAP, minor antenna proteins; MEP, methyl-erythritol phosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; NDH, NAD(P)H dehydrogenase; OEC, oxygen evolving center; PPDK, pyruvate orthophosphate dikinase; PPP, pentose phosphate pathway; PSI, photosystem I; PSII, photosystem II; RD, reductive phase; ROS, reactive oxygen species; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase. www.journal.frontiersin.org
CARBON FIXATION IN MESOPHYLL CELLS
During carbon fixation by mesophyll chloroplasts; first pyruvate is primed up to produce an energy rich 3 carbon compound called phosphoenol pyruvate. The enzyme responsible for this reaction is pyruvate di kinase which uses ATP and Pi for the priming
reactions; This is an unusual enzyme.
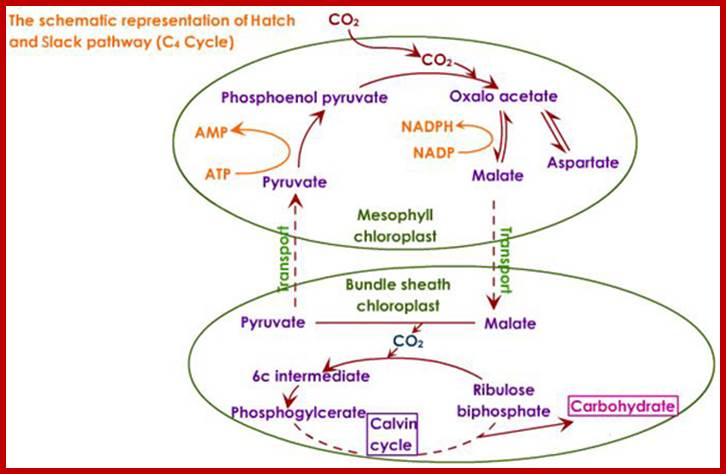
The phospho-enol pyruvate now acts as an acceptor of carbon dioxide and the product of this reaction is oxaloacetate. The enzyme called PEP carboxylase has a very low Km for CO2 and efficiently fixes carbon dioxide. In these reactions the first product of carbon fixation is a 4 carbon organic acid that is why these reactions are called C4 pathway. Chloroplasts are then transported across the cell walls into bundle sheath cells through the protoplasmic stands.
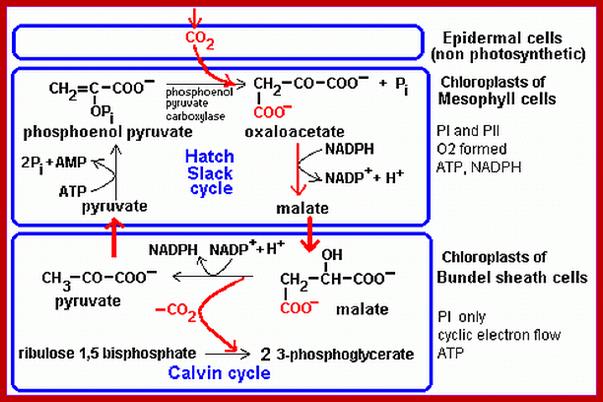
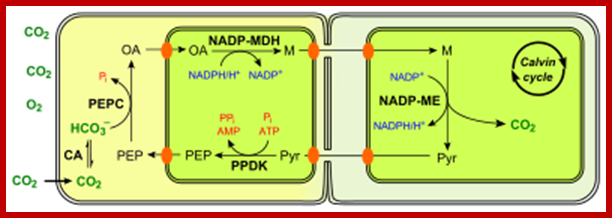
NADP-ME type of the C4 pathwaym.facebook.com; http://en.wikipedia.org/
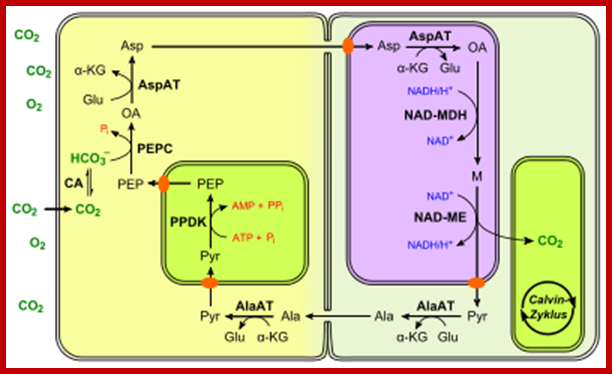
NAD-ME type of the C4 pathway; http://en.wikipedia.org/
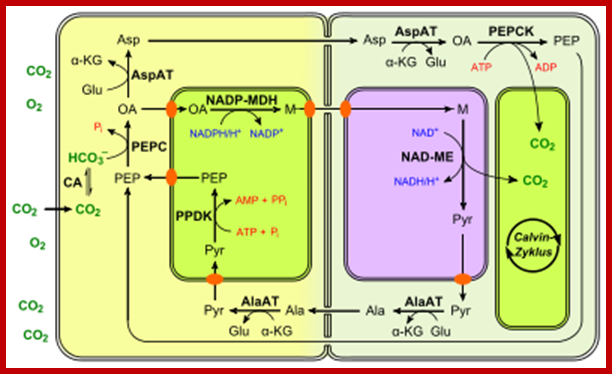
PEPCK type of the C4 pathway; http://en.wikipedia.org/
Carbon fixation in Bundle Sheath Cells
The chloroplasts found in bundle sheath cells use malate and the same is decarboxylated to pyruvate by malate decarboxylase enzyme. In this process a molecule of NADPH2 is formed by dehydrogenation reaction.
The CO2 released in this process is assimilated by RUBP with the help of RuBp carboxylase enzyme. The product of this reaction is, PGA which is then subjected to Calvin’s cycle process to produce a molecule of glucose as well as RUBP. And this process is repeated as long as carbon dioxide is available. The enzyme RUBP carboxylase present is also very efficient in carbon fixation.
Because of the low km action of carbon fixing enzyme C4 plants are considered as highly efficient in terms of net product of photosynthesis. The above plants are also tolerant to high temperatures and they are capable of fixing about 40-80 mg of C2 per dm2 of the leaf area per hour. In addition, as there plants contain very few micro bodies, they do not operate photo respiratory process, which is very common in C3 plants; thus they don’t lose any of its photosynthetic products.
Gene engineering: Plant bioengineers are working on converting C3 plants into C4 plants for C4 plants are efficient which can fix 50% more carbon than C3 plants. Scientist are working on Rice and other plants.
Crassulacian Acid Metabolic pathway
Plants belonging to Crassulacian family are mostly succulents. Such members are also found in other groups of plant kingdom. These plants possess large cells containing a number of chloroplasts. The cell sap has certain chemicals which prevent the loss of water from the surface of the plant body. Thus they prevent loss of water and survive even under drought conditions. Another interesting feature of CAM plants is the diurnal change in the opening and closing of the stomata parallel with the diurnal change in the organic acid content of the cell sap.
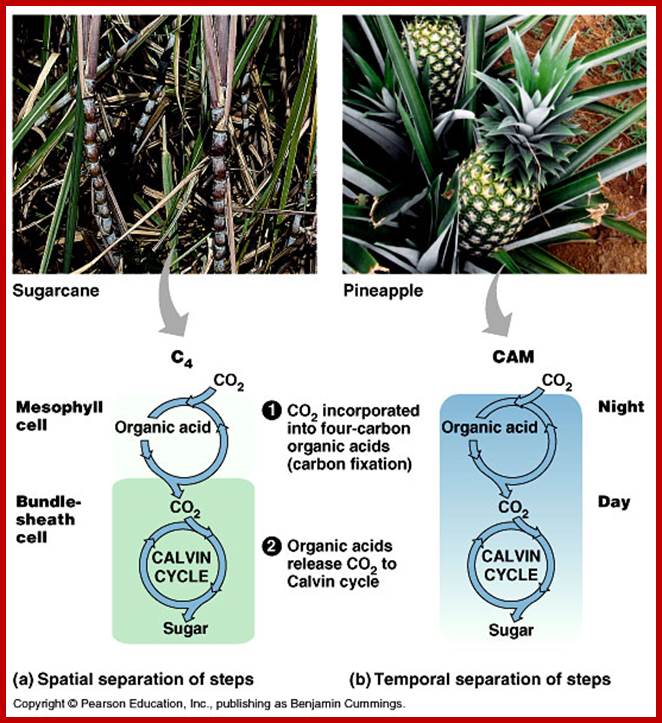
The Crassulacian plants exhibit closure of the stomata during day period and they open during night times. During night time, the cellular structures fix CO2 into malate to such an extent, the pH of the cell sap is rendered very acidic. But during the day period, the acidity of cell sap disappears because malate is decarboxylated and the products of these reactions are used for fixing carbon dioxide by C3 cycle.
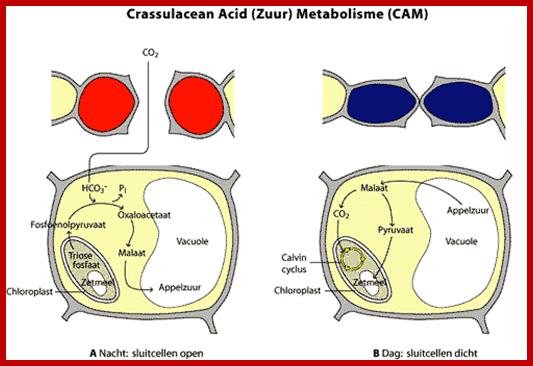
The CAM plants have developed certain structural and functional features which are well suited for fixing CO2 both at day and night periods. Though the chloroplasts do not show any structural dimorphism or compartmentalization, the switching on the one set of enzymes at daytimes and the other set at night is very remarkable. These plants fix CO2 efficiently under extreme conditions like drought, intense sun light wish temperature and poor availability of CO2. That is why these plants grow well in xerophytic conditions.
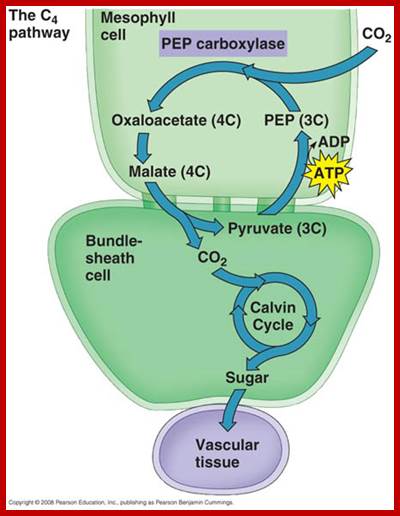
MECHANISM OF CARBON FIXATION IN SUCCULENTS;
Chloroplasts present in the cells of succulents do possess photosynthetic units as in any other plants. The light reactions and their products are same as that of C3 plants. But CAM plants, at night times, fix CO2 liberated due to respiration and at the same time, they also use atmospheric CO2 fix into malate.
The primary acceptor of CO2 in these plants is PEP, which by the action of PEP carboxylase fixes most of the available CO2 into OAA, which is then converted into malate by the activity of malate dehydrogenase. The required NADPH2 is supplied by Hexose monophosphate pathway. Thus, malate is continuously produced and stored in the cell sap during night times. Though the cell sap is highly acidic, it won’t inhibit normal cytoplasmic activities because of compartmentalization. Recent investigations suggest that at night times, RUBP also acts as the acceptor of CO2 and C3 cycle operates to some extent, as a result certain amount of glucose is also produced at night times. However, malate pathway is preponderant and predominant over C3 pathway.
Nonetheless, during day periods chloroplasts start fixing CO2 by normal C3 pathway. Meanwhile, the stored malate is decarboxylated and CO2 thus released is fixed by RUBP carboxylase into PGA. The same is subjected to reduction and regeneration steps of C3 pathway. Thus, Hexose is continuously synthesized. The remarkable feature of CAM plants is that they operate C3 and C4 pathway at different times; both operate sometimes, simultaneously.
Photorespiration:
It is also known as oxidative photosynthetic
carbon cycle or C2 photosynthesis. It tries t reduce the wasteful oxygenation
by the enzyme RuBisCo. RuBisco instead of fixing CO2 often adds oxygen the
product cannot be used in Calvin’s-Benson cycle. This reduces the Calvin’s
cycle ofCO2 fixation by 25% in C3 plants. Photorespiration is an interesting
process that exchanges metabolites between chloroplasts, leaf peroxisome and
mitochondria. Photorespiration takes place at the cost of One ATP and one
NA(P)H. This process also leads to the production H2O2 a dreadful oxidation
product which must immediately catalyzed by catalase. The conversion of 2x
2Carbon glycine to 1 C3serine in the
mitochondria by the enzyme glycine-decarboxylase is a key step, which releases
CO2, NH3, and reduces NAD to NADH. Thus, 1 CO2
molecule is produced for every 3 molecules of O2 (two deriving from
the activity of RuBisCO, the third from peroxisomal oxidations). The
assimilation of NH3 occurs via the GS-GOGAT cycle, at a cost
of one ATP and one NADPH.
Cyanobacteria have three possible pathways through which they can metabolize 2-phosphoglycolate. They are unable to grow if all three pathways are knocked out, despite having a carbon concentrating mechanism that should dramatically reduce the rate of photorespiration. http://en.wikipedia.org/
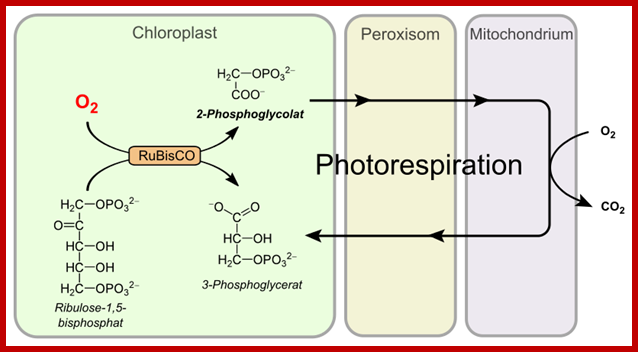
Simplified C2 cycle; http://en.wikipedia.org/
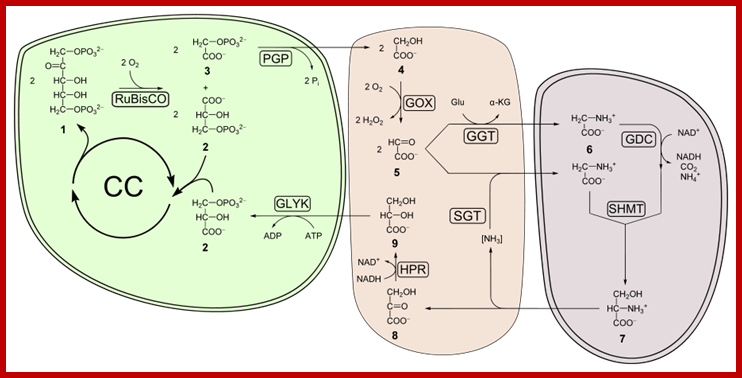
RuBp+O2 →
Phosphoglycolate + 3-phosphoglycerate + 2H+;www.aolanswers.com
FACTORS THAT CONTROL PHOTOSNNTHESIS
Law of limiting factors
Biochemical reactions are always controlled by one or more factors. The rate of reaction depends upon the availability of the required factors at optimal concentrations to achieve the maximal level. If any one of the factors is missing, then the rate of reactions suffers. Such factors are called as the limiting factors.
The optimal level of each of the factors can be determined by monitoring the rate of reaction by varying the concentration of the said factor while keeping the other factors more than needed for the maximal activity. When the rate of reaction is plotted against the concentration of the said factor a bell shaped curve is obtained. From this, it is possible to determine the optimal concentration at which the rate of reaction is at maximum. Nevertheless in many situations, even under optimal concentration of a particular factor, the rate of reactions remains constant. This is due to the deficiency of some other factor which controls the said reaction. If the concentration of deficient factor is increased, then the rate of reaction also increases, till other acts as a limiting factor. If such a factor is added the rate of reaction further increases. Thus in a multiple factor dependent reactions, all the said factors are required of the optimal level for the maximal rate of reaction. This concept of law of limiting factors was introduced by Balckmann. The factors that control the process of photosynthesis are Co2, pigments light, temperature, oxygen, water, genetic factors. Etc.
CARBON DIOXIDE
Global-CO2 concentration distribution; http://www.jpl.nasa.gov/
NASA, NOAA to Announce 2015 Global Temperatures, Climate Conditions;
;https://www.nasa.gov
During the course of evolution, heterotrophic organisms have acquired the ability to trap the solar energy into chemical energy and such auto tropic organisms were called plants and the same colonized the entire landscape of the earth. The successful story of photosynthetic plants is endowed with their genetic potentiality to produce light harvesting pigments, proteins and other enzymes to convert light energy into chemical energy and to fix CO2 into glucose. About 350 million years ago, the concentration of CO2 in the atmosphere was believed to be 20%. Since then the concentration ofCO2 is depleted and today the concentration is just 0.03 to 0.04% or less than that. Paradoxically increase in the concentration ofCO2 increases carbon fixation, so more vegetation, it can overcome or counteract with concept of danger of CO2.
The enzyme that is remarkable efficient in the fixation of CO2 is RUBP carboxylase, which is present in plants in large quantities. In fact, more than 50% of the total proteins found in leaves is RUBP carboxylase. On the other hand, the concentration of COP is so low; it acts as the limiting factor which affects the total output of photosynthate. It has been demonstrated that the increase in the concentration of CO2 up to 1% or little more, increases the rate of photosynthesis and its products.
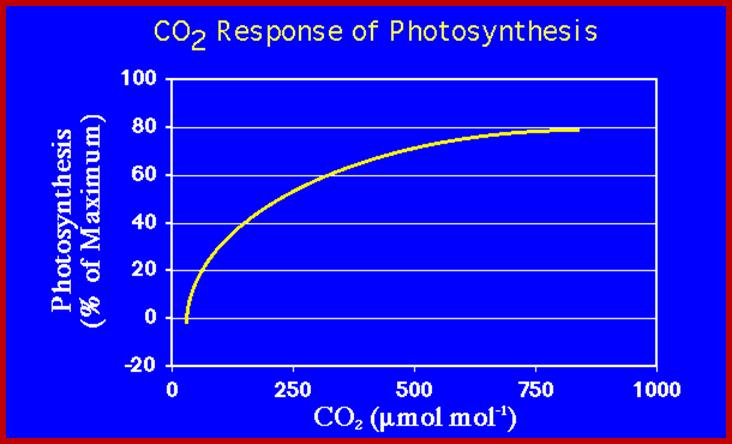
http://eesc.columbia.edu/
Calculations based on the total population of photosynthetic plants on this planet including plants growing in ocean and on land, the total amount of CO2 fixed amounts to about 175 billion tons per year, out of this only, 30-40% is consumed or utilized by biological activities. The rest is stored in the form of organic matter.
When CO2 emission by human activities is a tiny % of CO2 emissions why shout but take care emission is reduced. The oceans contain 37,400 billion tons (GT) of suspended carbon, but the land biomass has 2000-3000 GT. The atmosphere contains 720 billion tons of CO2 and Human contribute only 6GT. The oceans and land atmosphere exchange CO2 thus additional load by humans is incredibly small (Jeff Id); what Science says”
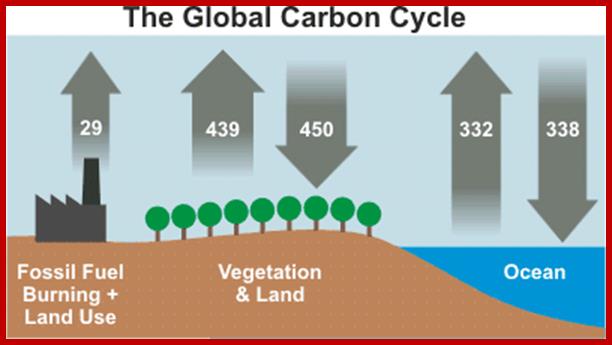
Global carbon cycle. Numbers represent flux of carbon dioxide in gigatons (Source: Figure 7.3, IPCC AR4). www.skepticalscience.com
But consider what happens when more CO2 is released from outside of the natural carbon cycle – by burning fossil fuels. Although our output of 29 gigatons of CO2 is tiny compared to the 750 gigatons moving through the carbon cycle each year, it adds up because the land and ocean cannot absorb all of the extra CO2. About 40% of this additional CO2 is absorbed. The rest remains in the atmosphere, and as a consequence, atmospheric CO2 is at its highest level in 15 to 20 million years (Tripati 2009). (A natural change of 100ppm normally takes 5,000 to 20,000 years. The recent increase of 100ppm has taken just 120 years). Making Sense of Climate Science Denial
PIGMENTS
Photosynthetically active plants contain various kinds of pigments of which Chl. A acts as the primary pigment and the others act as accessory pigments. Such pigments, in association with specific proteins, are capable of absorbing solar energy and the same is made available for other enzymes to convert it into chemical energy, which is ultimately stored in carbohydrates.
Though Chl. A pigments perform the primary process of photosynthesis, but accessory pigments too, absorb light energy and transfer the energy from molecule to molecule and finally to Chl.A. Apart from this function, the accessory pigments, particularly carotenoids protect the primary pigments from the bleaching effect of sunlight called solarization.
LIGHT
Almost all living organisms on this planet are depending upon the solar energy directly or indirectly, as the main source of energy, but plants are endowed with the potentiality of utilizing the solar energy directly. Light plays an important role, in initiating photochemical reactions, as well as activating the process of chlorophyll and other related protein synthesis. The effect of quality of light, quantity of light and intensity of light of photosynthesis and its products is very interesting.
QUALITY OF LIGHT
When light rays of fall on the surface of photo synthetically active parts, plastids absorb photons of the light and initiate photochemical reactions. Spectral analysis of sunlight and its effect on photosynthesis suggests that only visible part of solar electromagnetic radiation is used in photochemical reactions. Though blue, green, yellow, orange and red lights of the VIBGYOR spectrum are absorbed by the functional chloroplasts, the action spectrum is blue and red light. The other bands absorbed by the photosynthetic pigments is transferred to the primary pigments i.e. Chl. A.
INTENSITY OF LIGHT
The intensity of light is measured in terms of foot candles or lux. At low intensities of light, the rate of photosynthesis is slow. But with the increase in the intensity of red and blue lights the rate of photosynthesis also increases up to a certain point, but beyond 2000-2500 ft. candles of intensity, the rate of photosynthesis remains constant, which indicates the saturation level of light. Intense sunlight beyond 2500 ft. candles damages chlorophylls either by photo bleaching or by denaturing the enzymes due to resultant high temperature. However, green plants do not get bleached in spite of very high level of sunlight, because carotenoids protect the green pigments from photo oxidation. Intense light also activates photorespiration because of higher levels of oxygen available to plants. This has a greater impact on the total photosynthetic products in C3 plants, but not in C4 or CAM plants.
QUANTUM EFFICIENCY
Total amount of sunlight required either for the release of one mole of oxygen or for fixing one mole of CO2 into carbohydrates can be calculated by analyzing the requirement of energy for each step and the total amount of energy stored in one mole of glucose. The difference gives quantum efficiency.
It is possible to calculate what is the total number of quanta of light energy is required to produce the required ATPs and NADPH+H in non-cyclic photophosphorylation process, 4 mole quanta of light is required to produce 1 mole of NADPH+H and 1 mole of ATP. So for the synthesis of 12 moles of NADPH+H and 12 moles of ATP, the required light energy is 48 moles quanta of light. For the other 6 moles of ATP, six mole quanta of light energy is required for cyclic photophosphorylation to produce the same. All in all, 54 mole quanta of light energy is required 1 to fix 6 mole of CO2 into one mole of glucose. In fact, one mole quanta of red light possesses about 47.6 K.cals. Therefore 54 moles of CO2 is fixed into one mole of glucose. In fact, one mole quanta of red light possesses about 47.6 K.cals. Therefore 54 mole quanta of red light is equal to 54 x 47.6 = 2470 K.cals of energy. This is the amount of energy absorbed and used to produce one mole of glucose, but 1 mole of glucose stores just 696 K. cal of energy. So the quantum efficiency of photosynthesis to produce one mole of glucose is-
696 K.cals x 100 / 2470 K.cals = 28.1%
There calculations are exclusive of the energy required for the liberation of oxygen. For the liberation of every mole of oxygen, 4 mole quanta of light are required. If calculations are made to relate the liberation of 6 mole of oxygen 4 x 6 = 24 mole quanta of light and fixation of 6 mole of CO2 into 1 mole of glucose (54 mole quanta), the overall quantum efficiency will be 18.4%.
696 x 100 / 2470 + 1122 = 18.4%
Based on the above calculation it is suggested that all the plants put together store about 13 x 10 K.cals of sunlight energy while fixing 175 x 10 tons of CO2 per year.
TEMPERATURE
Any biochemical reaction that is mediated by enzyme is temperature dependent. With the increase in temperature the rate of reactions also increases. Certain photochemical processes in Hill’s reaction such as capturing of photons, transfer of excitons and the excitation of molecules, radiations in the form of fluorescence are temperature insensitive. But the synthesis of ATP by photophosphorylation, reduction of NADP to NADPH H and other carbon pathway reactions are temperature sensitive because they are enzyme dependent reactions. So for their maximal activities, they require an optimal temperature of 25-30 degree C, but some tropical grass members require 30-40 degree C as the optimal temperature. However, the temperature higher than this is deleterious because enzymes undergo denaturation and they lose their functional abilities.
OXYGEN
Oxygen is one of the products of photosynthetic reactions. The atmospheric concentration of oxygen is 18-20% and it is the product of photosynthetic activity of plants. If the concentration of oxygen increases intracellularly, the C3 plants resort to photorespiration, because the peroxisomal enzymes become active. At the same time, the RUBP carboxylase, which is responsible for carbon fixation, now it acts as oxygenase and starts breaking down RUBP. Thus the total yield of the photosynthate gets reduced. In certain plants it has been estimated that photorespiration oxidizes nearly 18-20% of the total photosynthate.
Another deleterious effect is that the green pigments in the presence of intense sunlight and oxygen get oxidize and damage the functionally active chlorophyll pigments. This process is called photo oxidation. But C4 plants by virtue of not having micro bodies tolerate excess oxygen and do not suffer from it.
WATER
Water is an important substrate for photosynthesis, for it provides hydrogen ions for the production of NADPH+H. In addition, it is also used in many biochemical pathways during carbon fixation. The concentration of water in the plant body is very important because deficiency of it causes closing of the stomata, and also it affects various metabolic activities, hence the rate of photosynthesis is also affected.
GENETIC FACTORS
Whatever may be the optimal levels of required factors available for the plant, it is ultimately the genetic factors that control the efficiency of plants potentiality to perform photosynthesis. For example, C4 plants because of their dimorphic chloroplasts, they are capable of fixing CO2 more efficiently than C3 plants. Similarly, CAM plants are capable of fixing CO2 both at night and day times.
The number of chloroplasts, structural adaptation of chloroplasts, functional efficiency of chloroplasts, the number of foliages, orientation of foliage and the overall cellular enzymatic features. The high yield of plants in terms of photosynthesis is ultimately controlled by the potentiality of plant genome.
PHOTORESPIRATION
Besides the oxidation of glucose into CO2 and H2O by regular respiratory process, many dicot plants with C3 characters oxidize pentose sugars in the presence of light higher concentration oxygen by a process called photorespiration. It differs from dark respiration in being insensitive to cyanide inhibition.
Photorespiration is due to the activity of micro bodies found in mesophyll cells of C3 plants. In mesophyll cells, micro bodies are closely associated with chloroplasts on one side and mitochondria at the other. On the contrary, C4 plants do not possess any micro bodies, even if present; there will be only a few. Hence, C4 plants do not oxidize their photosynthetic products but C3 plants loose about 18% or more of their total photosynthate by photorespiration. That is why C4 plants are considered as efficient when compared to that of C3 plants.
MECHANISM OF PHOTORESPIRATION
On a bright day, when the process of photosynthesis is at its peak, the concentration of CO2 goes down from 0.03% to 0.005% or so. At the same time, the concentration of oxygen increases considerably.
In the presence of higher amounts of oxygen, RUBP carboxylase acts as RUBP oxygenase. Normally RUBP carboxylase fixes carbon dioxide, but in these conditions of high O2, RUBP carboxylase exhibits oxygenase activity. This form of enzyme oxidizes RUBP molecules into phosphoglycerate and phosphor-glycollate. For this oxidation reaction the enzyme uses the available oxygen. This process takes place in functional chloroplasts. The phosphor-glycollate is then dephosphorylated to glycol late by a phosphatase enzyme.
The glycollate is then transported across the membrane to peroxisomes which are very closely associated with chloroplast membranes. In peroxisomes, glycol late is acted upon by glycollate oxidase to produce glyoxylate and H2O2. A molecule of oxygen is utilized in this process. However, the H2O2 produced in this reaction is immediately catalyzed by peroxidase enzymes to liberate H2O and O2. Then the glyoxylate is converted to glycine by transaminase reactions.
Glycine produced in the peroxisomes is not further metabolized. Glycine is then transported across the membranes into mitochondria which is also associated with peroxisomes. In mitochondria, two molecules of glycine are condensed to a 3 carbon serine by a series of decarboxylation and deamination reactions.
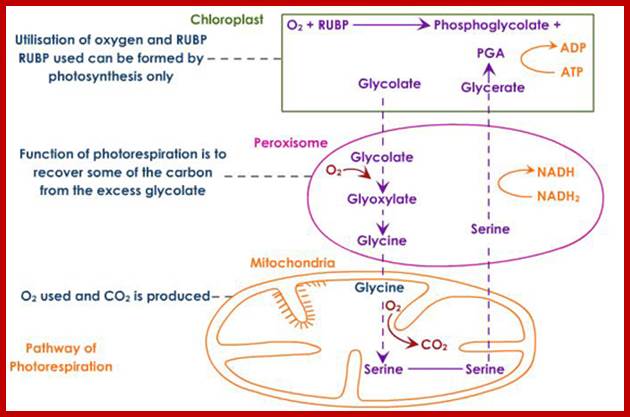
fig. below www.uic.edu
Below figure shows; Interactivitivities beween Plastids and Mitochondria:.
![]()