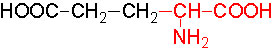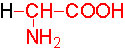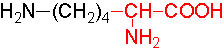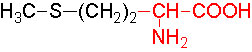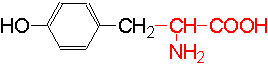Nitrogen Metabolism
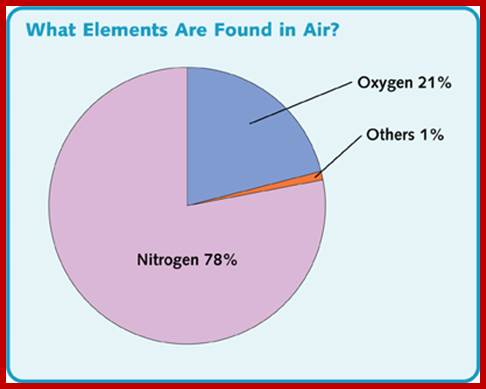
Nitrogen is a very important constituent of cellular components. Alkaloids, amides, amino acids, proteins, DNA, RNA, enzymes, vitamins, hormones and many other cellular compounds contain nitrogen as one of the elements. It is not exaggerating to say that Nitrogen is the key element for it is the most important constituent of proteins and nucleic acids. Thus, N2 plays a significant role in the formation of the above said compounds which in turn control cellular activities. Without nitrogen, no living organism can survive. Paradoxically all the living organisms are virtually submerged in a sea of atmospheric nitrogen (i.e. 78%), but unfortunately not all organisms are endowed with the potentiality to utilize this abundantly available molecular N2 directly. Only some organisms like certain bacteria, blue green algae and few fungi, have the potentiality to utilize molecular N2 directly and fix it. However, most of the plants are capable of utilizing other forms of nitrogen with ease and facility.
Let us have look at major metabolic pathways in a figure that shows most but not all.
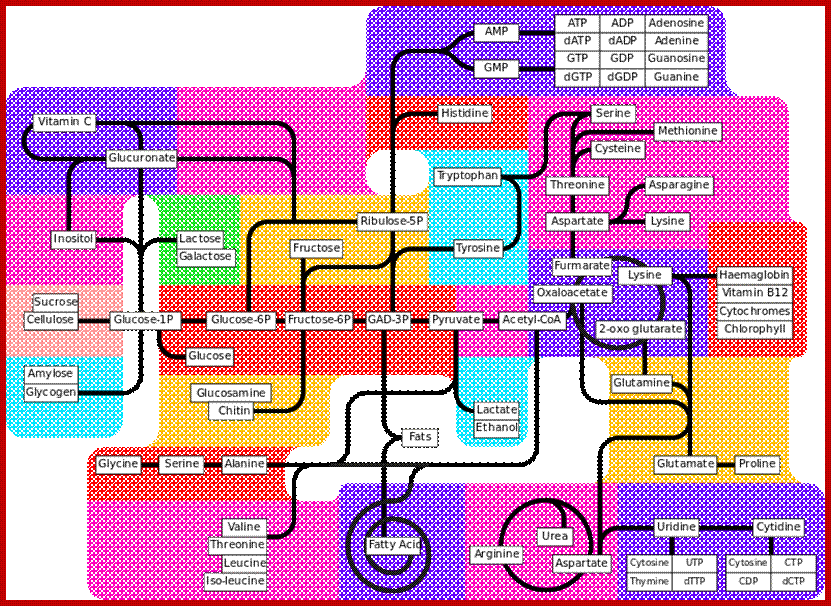
Major Metabolic Pathways; https://en.wikipedia.org
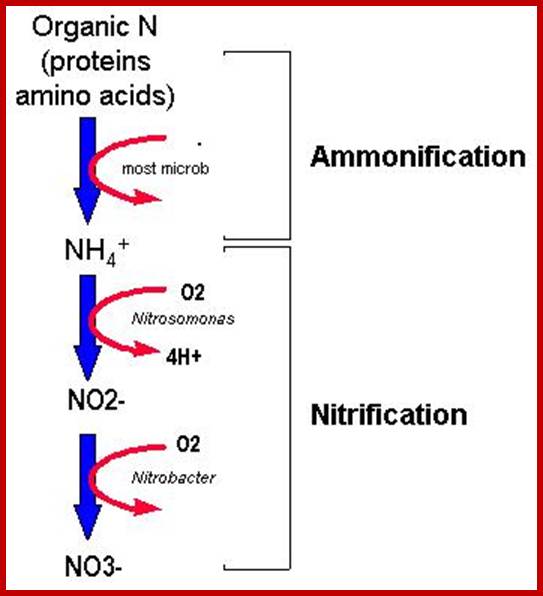
Nitrogen enters roots as NO3- or NH4+. Nitrate reduction results in the production of NH4 called N2 fixation. NH4 is 1st incorporated into amino acids via the glutamine synthetase (GS) reaction. The NH4+ is incorporated into amino acids in roots and leaves and the amino acids get integrated into proteins. The main if not sole function of some proteins is to provide a store of amino acids (see below).
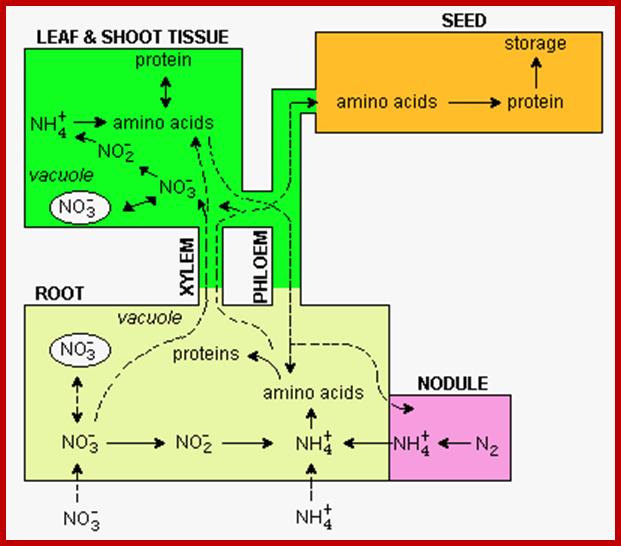
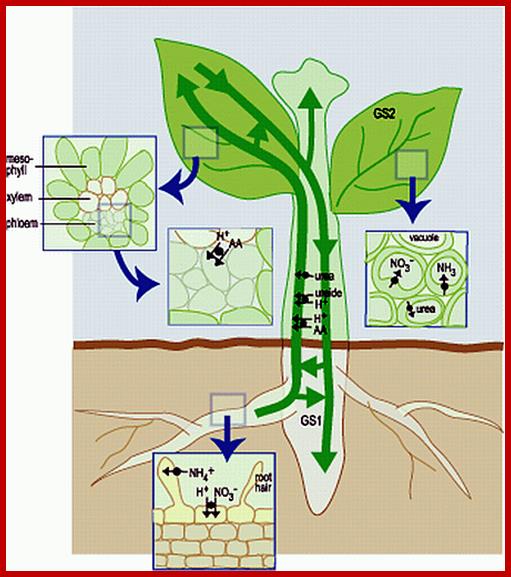
Absorption and Transport of minerals, NH4 and Nitrates by roots; cell biol; http://slideplayer.com
SOURCES OF NITROGEN
Elemental Nitrogen, Ammoniacal and organic form of Nitrogen:
Ammonical form of N2 is available in soil in the form of urea or NH4 in free-state. Urea, if present, is first split into NH4 and CO2, and NH4 is then utilized directly by metabolic pathways by higher plants. But recent studies indicate that urea can be directly used up by metabolic pathways in certain plants. It should be remembered here, that free ammonia is the only utilizable form of N2 that can be directly incorporated into amino acids. Whatever may be the source of nitrogen, first it has to be converted to NH3 and fixed into amino acid. It can be converted or transferred to other forms by various pathways that operate in living systems.
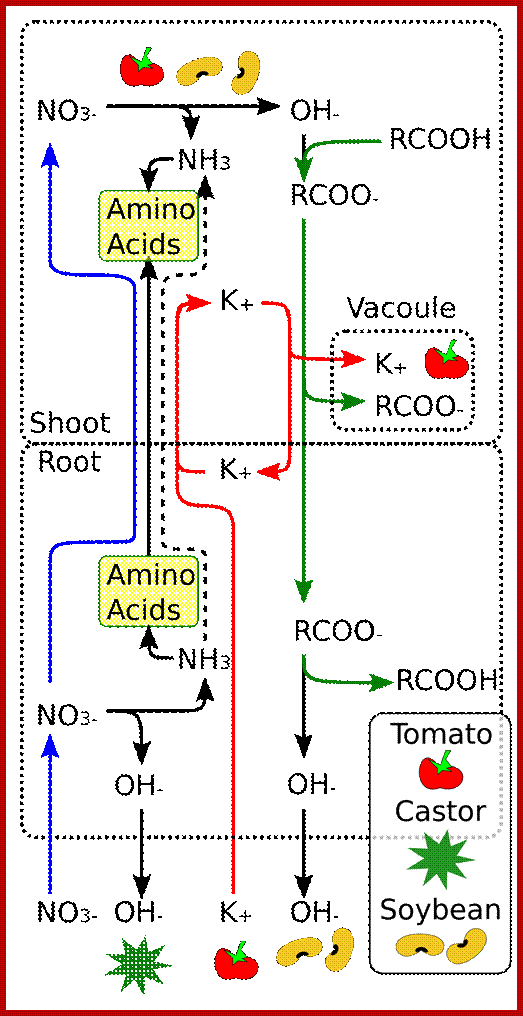
Different plants use different pathways to different levels. Tomatoes take in a lot of K+ and accumulate salts in their vacuoles, castor reduces nitrate in the roots to a large extent and excretes the resulting alkali. Soya bean plants move a large amount of malate to the roots where they convert it to alkali while the potassium recirculates; www.en.wikipedia.org
The decay of dead plants and animals also releases different kinds of nitrogen compounds of which amino acids, nucleotides (including DNA?) and other such Nitrate compounds constitute organic form of N2. The same are absorbed by the root system and utilized directly. Thus the decaying organic matter acts as the rich source of organic nitrogen that can be utilized by not only higher plants but also by micro-organisms.
NITRATE / NITRITE FORM
Invariably the N2 that is available in the soil is in the form of nitrates. And nitrites are also found but in small quantities. These forms are available as ions and the same are easily absorbed by the roots or cellular surfaces from its surrounding soil solution. The absorption of NO3 or NO2 ions is not by just diffusion process, but it is facilitated by specific carriers.
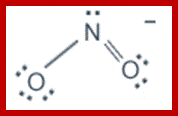
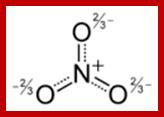
Nitrite and nitrate ions; www.wiredchemist.com
Once the nitrate or nitrite ions enter into cellular milieu they have to be converted to NH4, before the same can be incorporated into cellular components. Under normal conditions, nitrite is never accumulated in the soil in sufficient quantities and it is toxic to plants and to other microbes.
THE MECHANISM OF CONVERSION OF NO3 AND NO2 TO NH4
Plant structures like roots as well as leaves can utilize nitrates and the same can be converted to NH4. But more of nitrate reductive activity is found in leaves than in roots. However, the mechanism of nitrate and nitrite reduction is performed by different enzymes while NO3 is reduced by nitrate reductase enzymes and the NO2 is reduced by nitrite reductases.
NITRATE REDUCTION
Nitrate reduction to NH4 is not a single step process, but it is a series of reactions in which the first step is performed by nitrate reductase. This enzyme has been isolated and purified from various sources like Aspergillus, bacteria, chlorella, blue green algae, alfa alfa and other higher plants. The mol. Wt. of it is about 3.5 x 10^5 daltons. The enzyme is associated with 2 cofactors i.e., FAD and two molybdenum ions. The enzyme also requires reducing power supplied by NADH+H or NADPH+H. The former is available in non chlorophyllous tissues and the latter is found in chloroplast containing leaves.
NITRATE REDUCTASE IS AN INDUCIBLE ENZYME
In the absence of NO3 the amount of this enzyme present in the tissues is very low. With the addition of NO3 as the substrate, the amount of this enzyme increases many fold. However, the induction requires light without which the enzyme induction is not possible to the fullest extent. The nitrate induced enzyme synthesis can be inhibited by the inhibitors of transcription and translation like actinomycin D and cycloheximide respectively, which indicates that NO3 acts as an inducer of nitrate reductase gene expression. How light modulates the gene expression is not yet clear.
Furthermore, phytohormones, particularly cytokinin also induces nitrate reductase synthesis denovo even in the absence of light and NO3. Cytokinin induced NO3 reductase activity can be inhibited with actinomycin or CHI. The mechanism of denovo synthesis of nitrate reductase, though not clear, it is fully accepted that the nitrate reductase is an inducible enzyme.
NITRATE REDUCTION
The reduction of nitrate to ammonia is a multistep reaction in which nitrates are reduced to nitrites, which are then converted to hyponitrites then to hydroxylamines and finally to ammonia.
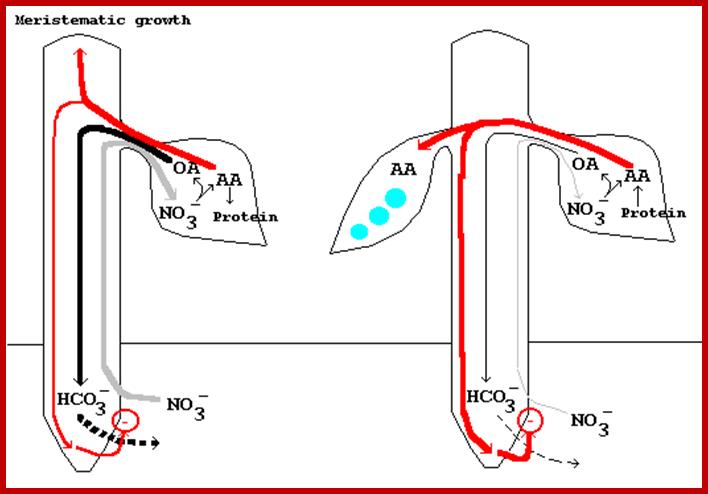
A model for the control of nitrate uptake by leaf-generated signals during rapid vegetative growth (left) and rapid pod fill (right) (Ismande and Touraine, 1994). During vegetative growth (left), nitrate ions are rapidly absorbed by the root and transported via the xylem to the leaf. In the leaf, nitrate reduction produces organic acids (OA) and amino acids (AA). Most of the newly formed OA are translocated to the
root where a carboxyl group is released in exchange for a nitrate ion, whereas the newly assimilated N is incorporated primarily into leaf N compounds (protein) (Ismande and Touraine, 1994). http://www.hort.purdue.edu/
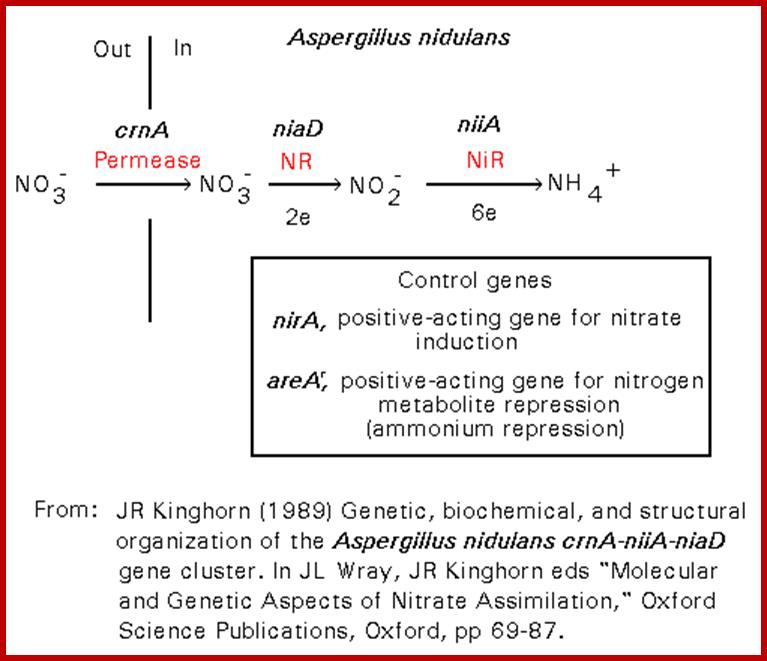
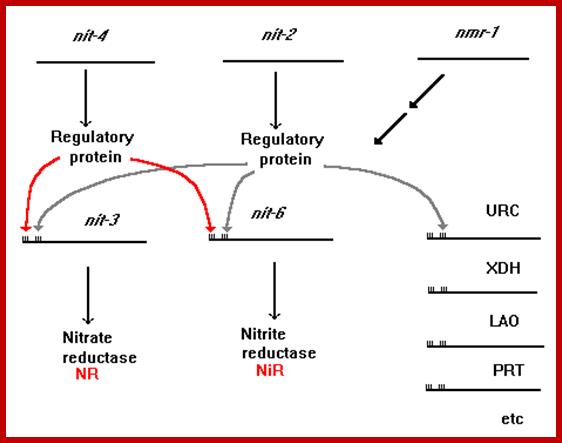
URC = Uricase; XDH = Xanthine dehydrogenase; LAO = L-Amino acid oxidase; PRT = Protease. The positive-acting major nitrogen regulatory gene, nit-2, and the pathway-specific regulatory gene, nit-4, are each presumed to encode a regulatory protein which is required for the expression of nit-3 (encoding NR) and nit-6 (encoding NiR).
The nitrogen regulatory circuit of N. crassa. Expression of genes that encode nitrogen metabolic enzymes only occurs upon nitrogen catabolite derepression and simultaneous induction by a pathway-specific metabolite. The repressing metabolite appears to be glutamine, although the nature of the factor with which it interacts is unknown. NIT-2 is a sequence-specific DNA-binding protein with a single zinc finger that acts globally to activate the expression of many structural genes that encode enzymes of nitrogen metabolism. NMR is a negative-acting regulatory protein that is required to establish nitrogen repression, and it appears to act by binding directly to NIT-2 to inhibit its function. NIT-4 is a pathway-specific positive-acting regulatory factor that has a zinc cluster DNA-binding motif. NIT-4 interacts with NIT-2 to turn on the expression of nit-3 and nit-6. Other specific positive-acting factors are required to express genes encoding enzymes for other nitrogen pathways. URC signifies uricase, for which the structural gene has not yet been identified in Neurospora. uc-5 and ud-1 also are subject to general nitrogen metabolite regulation. Original figure from; G. A. Marzluf. http://www.fgsc.net
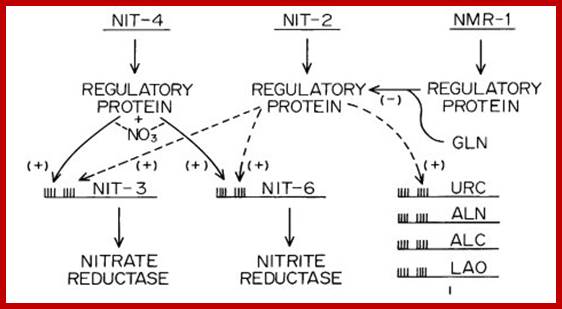
Two models for operation of the nitrogen control circuit are shown below. (a) The nit-2 gene is postulated to be expressed constitutively to yield a regulatory product active for turning on nit-3 and nit-6 unless it is inhibited by the metabolite repressor molecule, glutamine. (b) The nmr gene is visualized as producing a repressor protein which is activated by glutamine and precludes nit-2 gene expression. In the absence of glutamine, nit-2 is expressed and in turn activates nit-3 and nit-6 (Marzluf and Fu, 1989). Nitrate reductase is expressed in a constitutive fashion in nmr mutant strains, which appear to be largely insensitive to nitrogen catabolite repression (Fu et al, 1988). nmr mutants are sensitive to chlorate in the presence of ammonia or glutamine, whereas the wild type is chlorate resistant under these conditions (Fu et al, 1988). http://www.hort.purdue.edu/
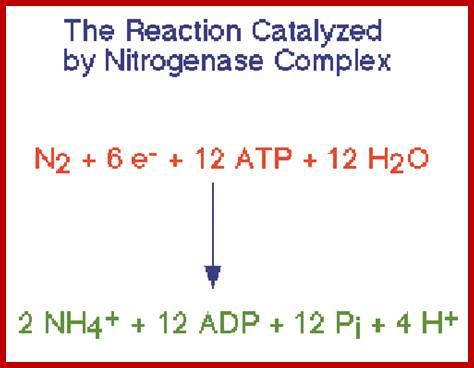
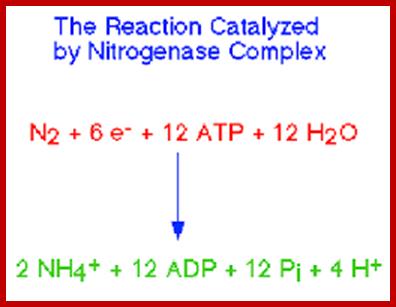
Top fig; Nitrogenase performs the fixation reactions, (Fig. 9) a six-electron reduction of N2 to NH3, using 6 NADPH and as many as 12 ATP. The nitrogenase complex can be irreversibly inactivated by atmospheric oxygen (O2). The sensitivity of nitrogenase to oxygen may be a remnant of earlier times, billions of years ago, when the Earth's atmosphere was lower in oxygen. Many of the organisms that can fix dinitrogen can only do so in environmental niches where oxygen is either absent or at very low levels. https://www.bio.purdue.edu
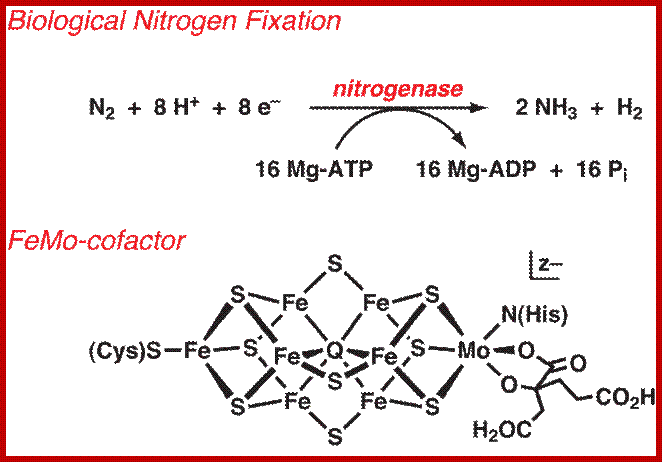
Bottom fig. Nitrogenase: A Molecular Inorganic Perspective
The nitrogenase enzymes fix atmospheric dinitrogen through the action of a singular class of metallocluster cofactor. For the most studied, most active enzyme subclass, this cofactor has been formulated as a structurally complex [MoFe7S9Q] cluster (the iron-molybdenum cofactor, FeMo-cofactor), where Q is a recently discovered interstitial monoatomic ligand believed to be nitride. http://www.science.uwaterloo.ca/
 Nitrogen fixation can be
carried out by certain organisms, primarily bacteria. These organisms produce
the enzyme nitrogenase,
which catalyzes the conversion of atmospheric nitrogen to ammonium. Some nitrogen fixing microbes can
carry out this process on their own, while others fix nitrogen in symbioses
with plants (symbiotic nitrogen fixation):;Nitrogen\fixation;
Anabaena, Rhizobia and Azolla, http://www.yorku.ca/
Nitrogen fixation can be
carried out by certain organisms, primarily bacteria. These organisms produce
the enzyme nitrogenase,
which catalyzes the conversion of atmospheric nitrogen to ammonium. Some nitrogen fixing microbes can
carry out this process on their own, while others fix nitrogen in symbioses
with plants (symbiotic nitrogen fixation):;Nitrogen\fixation;
Anabaena, Rhizobia and Azolla, http://www.yorku.ca/ 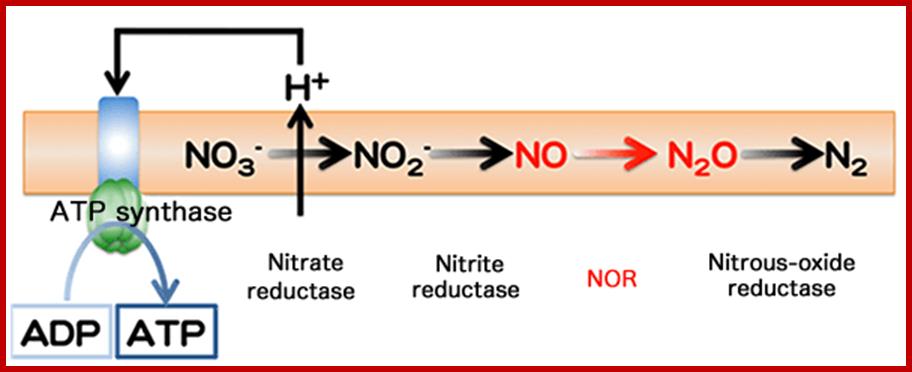
Adenosine triphosphate (ATP) production by denitrification and nitrate respiration
In denitrification, nitrogen oxides (NO3- and NO2-) are sequentially reduced to finally become N2 gas. During this process, NO and N2O are produced. Each step denoted by an arrow → is catalyzed by different enzymes. Membrane-protein nitrate reductase induces a concentration gradient of hydrogen ions through a cell membrane. Using this concentration gradient, ATP synthase catalyzed the synthesis of ATP from adenosine diphosphate (ADP) (nitrate respiration). http://www.spring8.or.jp/
Depending upon the tissues involved nitrate reductase accepts NADH2 (roots) or HADPH+H (leaves), where hydrogen is transferred to the coenzyme FAD to form FADH2. In the next step, protons (H+) and electrons are transferred to NO3 simultaneously. However, electrons are transferred to NO3 through molybdenum ions.
For the maximal activity of nitrate reductase, it requires an optimal concentration of MO2+, Fe3+ and Ca2+ ions. Though calcium has no catalytic activity in this enzymatic reaction, unlike iron and molybdenum which are involved in electron transport and it facilitates the transport of nitrite across the chloroplast membranes. Thus, the nitrite synthesized in this reductive step in the cytoplasm is transported into chloroplasts. But in roots, lower fungi and bacteria, the entire process takes place in the cytoplasm.

Nitrate and Periplasmic nitrate reduction; http://pubs.rsc.org/
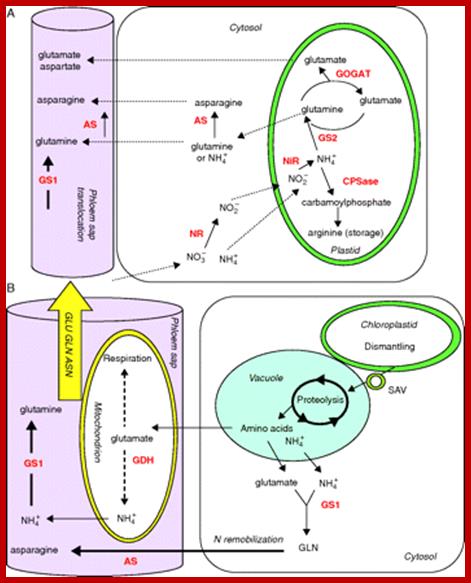
Schematic presentation of key enzymes involved in nitrogen management in (A) young and (B) senescing leaves. (A) Nitrate reductase (NR) and asparagine synthetase (AS) are localized in the cytosol, and nitrite reductase (NiR), glutamine synthetase 2 isoenzyme (GS2), glutamate synthase (GOGAT) and carbamoyl phosphate synthetase (CPSase) within the plastids of mesophyll cells. Glutamine synthetase isoenzyme 1 (GS1) and AS are located in the cytosol of companion cells. (B) Senescence-associated events include chloroplast degradation and translocation of plastid proteins to the central vacuole via senescence-associated vacuole (SAV) trafficking. Amino acid recycling occurred in mitochondria and cytosol of mesophyll cells and companion cells. Glutamate dehydrogenase (GDH), GS1 and AS are the major enzymes involved in the synthesis of glutamine, glutamate and asparagine in the phloem.; Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture; http://aob.oxfordjournals.org/
NITRITE REDUCTION
In most of the higher plants so far studied, the nitrites synthesized in cytoplasm are transported into plastids, where the nitrites are reduced to hyponitrite by an enzyme called nitrite reductase. The enzyme has a mol. Wt. of 60-70KD and it has a special heme component called siroheme detected in soret band. Actually, there are two forms of nitrite reductases, of which one form uses NADPH+H as the proton/electron donor in photosynthetic tissues, but root tissues and others including bacteria and fungi use NADH+H as the hydrogen donors. The enzyme nitrite reductase possesses flavin and iron groups. Added to this, they are inducible enzymes. Strangely, these enzymes are induced by nitrates than nitrites. However, nitrite reductase brings about the reduction of nitrite to NH4 in a multistep reaction, where the intermediary products remain attached to the surface of enzyme; only the final product is release from the surface. In this process, a total of six electrons and six protons are transferred to nitrite to produce ammonia.
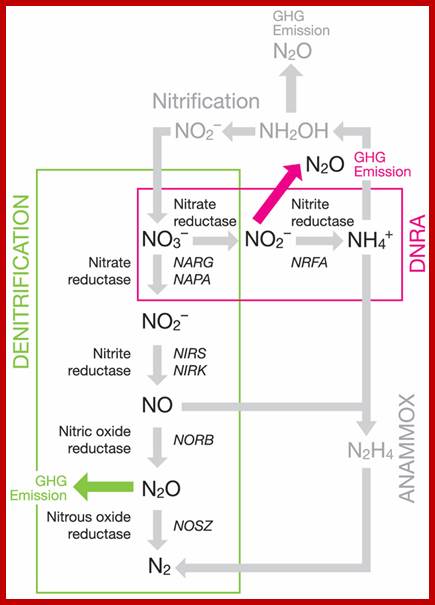
Partial soil nitrogen cycling pathways with an emphasis on denitrification and DNRA showing the enzymes involved and genes commonly used as markers; http://www.frontiersin.org/
Nonetheless, in some cases one of the intermediate products like hydroxylamine has been found to be converted to NH4 by the activity of hydroxylamine reductase. Such reactions have been observed in mesophyll tissues of higher plants, Neurospora, aspergillus and some bacteria. Whether or not, the enzyme nitrite reductase by itself is capable of converting hydroxylamine to NH4 is not clear. Still the overall pathway from NO3 or NO2 to NH4 is catalyzed by a group of enzymes or multienzyme complexes, but the synthesis of NH4 is very essential for amino acid synthesis.
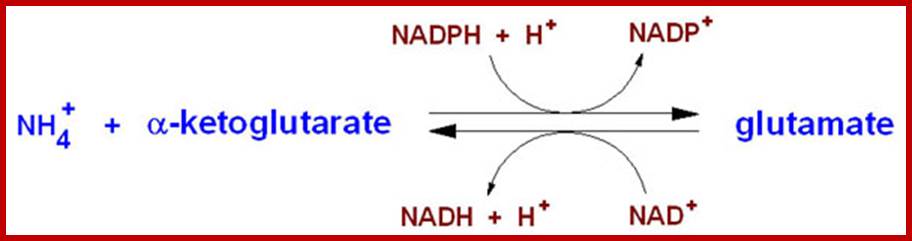
http://themedicalbiochemistrypage.org/
The glutamate dehydrogenase utilizes both nicotinamide nucleotide cofactors; NAD+ in the direction of nitrogen liberation and NADP+ for nitrogen incorporation. In the forward reaction as shown above glutamate dehydrogenase is important in converting free ammonia and α-KG to glutamate, forming one of the 20 amino acids required for protein synthesis. However, it should be recognized that the reverse reaction is a key anapleurotic process linking amino acid metabolism with TCA cycle activity. In the reverse reaction, glutamate dehydrogenase provides an oxidizable carbon source used for the production of energy as well as a reduced electron carrier, NADH. As expected for a branch point enzyme with an important link to energy metabolism, glutamate dehydrogenase is regulated by the cell energy charge. ATP and GTP are positive allosteric effectors of the formation of glutamate, whereas ADP and GDP are positive allosteric effectors of the reverse reaction. Thus, when the level of ATP is high, conversion of glutamate to α-KG and other TCA cycle intermediates is limited; when the cellular energy charge is low, glutamate is converted to ammonia and oxidizable TCA cycle intermediates. Glutamate is also a principal amino donor to other amino acids in subsequent transamination reactions. The multiple roles of glutamate in nitrogen balance make it a gateway between free ammonia and the amino groups of most amino acids.
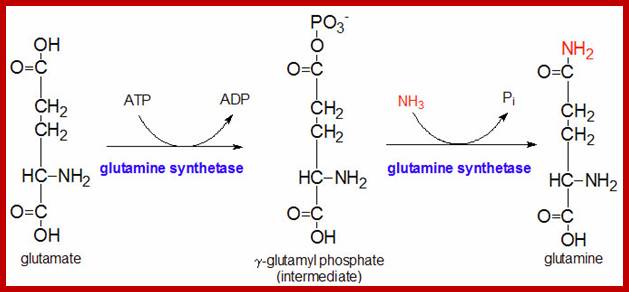
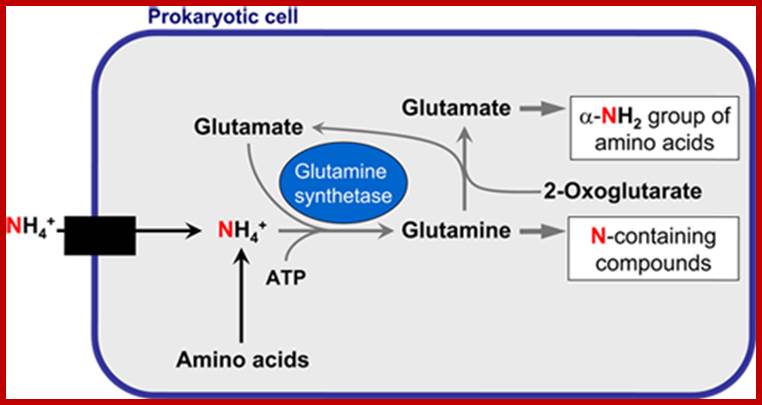
The glutamine synthetase reaction is also important in several respects. First it produces glutamine, one of the 20 major amino acids. Second, in animals, glutamine is the major amino acid found in the circulatory system. Its role there is to carry ammonia to and from various tissues but principally from peripheral tissues to the kidney, where the amide nitrogen is hydrolyzed by the enzyme glutaminase (reaction below); this process regenerates glutamate and free ammonium ion, which in humans is excreted in the urine.;www.themedicalbiochemistrypage.org
www.themedicalbiochemistrypage.org
www.pecinta-kimia.blogspot.com
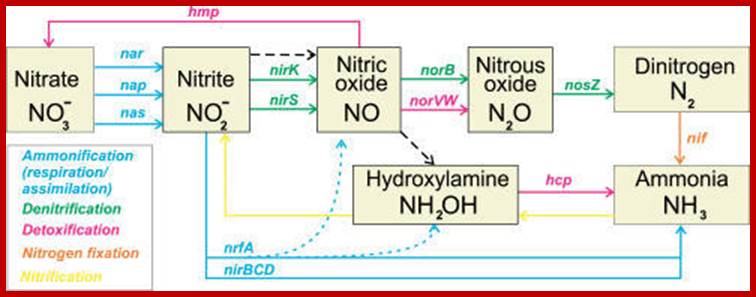
The Bacterial Inorganic Nitrogen Cycle; The ammonification, denitrification, detoxification, nitrogen fixation, and nitrification pathways are shown by colored solid lines with genes names involved in the pathway. The dashed black line shows possible non-enzymatic interconversions of nitrogen oxides. The dotted line shows additional formation of NO and hydroxylamine during nitrite ammonification; http://openi.nlm.nih.gov/
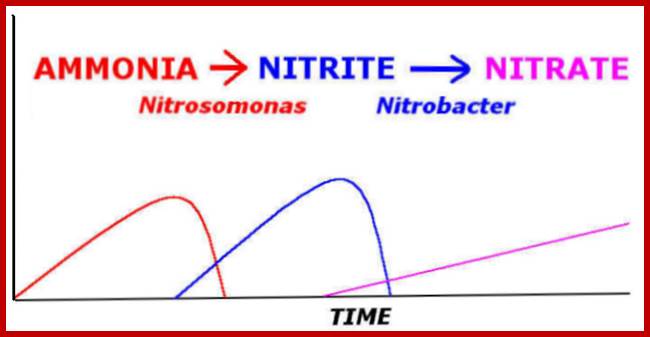
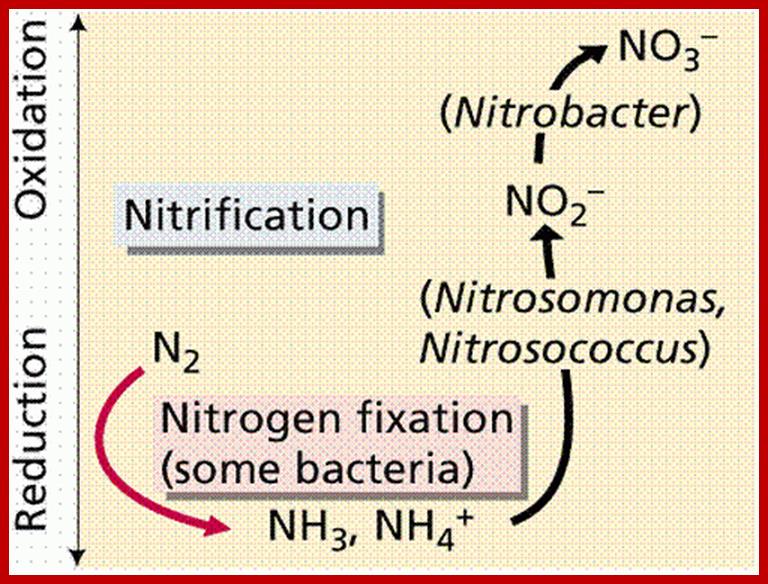
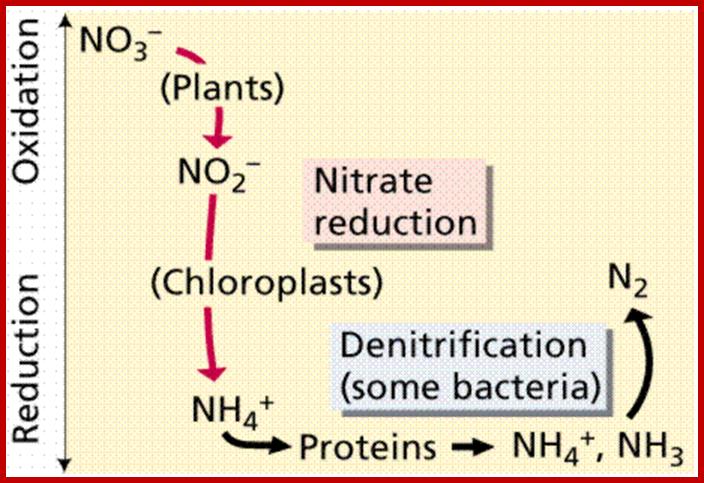
imgarcade.com; http://www2.estrellamountain.edu/
Nitrosomonas; http://imgarcade.com/
MOLECULAR NITROGEN
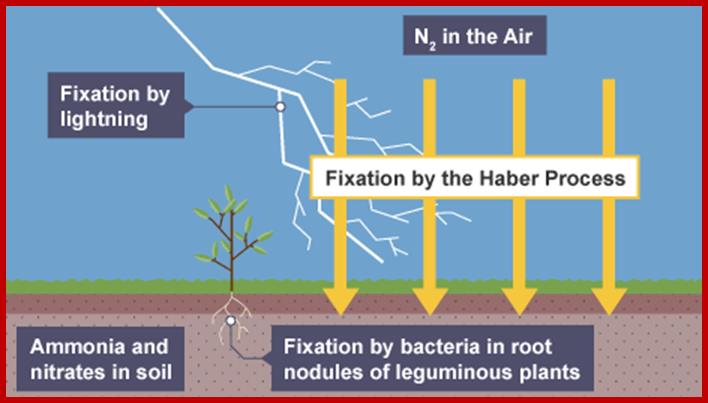
Abundantly available molecular N2 is more or less inert. With the exception of some bacteria, fungi and blue green algae none of the higher plants are capable of utilizing molecular N2 directly. However, nature has devised mechanisms to fix this type of N2 into utilizable form of N2 i.e., NH4 by non-biological and biological methods.
http://science.jrank.org/
Fixation of Inorganic N2:
Atmospheric N2 is converted into Ammonia by Haber’s process; then ammonia is converted into different forms of nitrates, which is used by bacteria and plants.
N2 + 8H+ + 8e- + 16ATP = 2NH3 + H2 + 16ADP = 16 Pi
NITROGEN FIXATION
Non biological Method:
Electrical discharges in atmosphere due to lightening leads to the formation of various oxides and reductants of N2. In the presence of water vapors they dissolve and produce nitrous and nitric acids. These, in turn, come down to earth along with rain water. Later they get converted to nitrates. Annually many billion tons of atmospheric N2 is fixed by this non biological process.
BIOLOGICAL METHOD – ASYMBIOTIC PROCESS
Among the living plant world, some free-living bacteria, fungi and blue green algae are capable of fixing molecular nitrogen into utilizable form of N2 i.e., NH4. Ex. Azatobactor veinlandi, Clostridium pasteurianum, Rhodospirullum rubrum, Chromatium, Nostoc, Anabaena, Rivularia, etc. When the above said organisms are allowed to multiply in the soil, under favourable conditions they easily fix 15-40 kgs. of N2 per acre per year. In recent years, the above said organisms are made available to farmers as bio-fertilizers.
When the cultures of them are spread in the fields and allowed to grow, they enrich the soil with a lot of nitrogen as a natural fertilizer. The mechanism by which molecular N2 is converted to NH4 is described elsewhere. One important aspect of it is to maintain moisture in the soil. This living fertilizer renewable and enriches the soil all the time.
SYMBIOTIC PROCESS
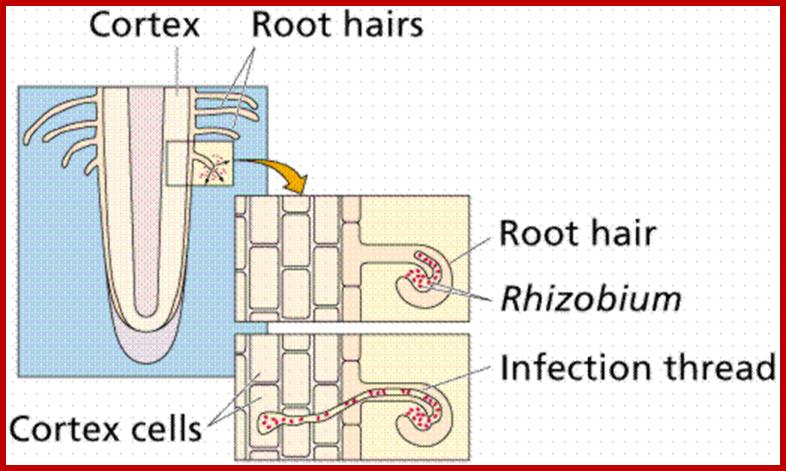
None of the known crop plants, or any other angiosperms are capable of utilizing molecular N2 directly, but some have developed a method by which they obtain nitrogen through symbiotic association with bacteria. It is widely known that many species of bacteria and also some blue green algal colonies live in association with higher plants, either in the roots, leaves, lichens, liverworts and coralloid roots. But the roots of leguminous plants possess characteristic root nodules in which nitrogen fixing bacteria called Rhizobium are present. These bacteria, on infecting host roots induce the development of characteristic pink colored root nodules. In their symbiotic association, bacteria obtain carbohydrates and other minerals from host cells and host cells in return obtain nitrogen fixed by bacteria. So by growing leguminous plants in the fields the soil will be enriched with nitrogen fertilizers up to the tune of 40-80 kg./acre./year.
DEVELOPMENT OF ROOT NODULES
SPECIFICITY OF BACTERIA AND HOST ASSOCIATION
The symbiotic association between bacteria and the host is highly specific. For example, Rhizobium phaseolin infects phaseolus species only but not others. Similarly, Rhizobium trifoli infects Trifolium repens but not others. The host bacterial specificity is due to the presence of glycoproteins as receptors in host root cell surface which recognizes some proteins found on the bacteria cell wall. These recognize each and other as in the case of enzyme recognizing its specific substrate.

Symbiotic Remorins; http://www.genetik.bio.lmu.de/
Legumes have the unique ability to undergo symbiotic associations with bacteria belonging to the Rhizobiaceae family. These Rhizobacteria secrete signaling molecules (Nodule Factors) that trigger physiological and morphological plant responses. In the course of this interaction, rhizobia invade the host root leading to the formation of a novel symbiotic plant organ: the root nodule. Rhizobia remain surrounded by a plant-derived plasma membrane from the very first moment of host invasion until age-dependent degradation of the symbiotically active bacteroids. This membrane serves as an essential interface for plant-microbe signal transduction and is certainly one of the key determinants for the success of the association. Using DNA microarrays, a remorin gene (SYMREM1) that is strongly induced in root nodules of Lotus japonicus and Medicago truncatula (Colebatch et al., 2004; El Yahyaoui et al., 2004) has been identified. genetik.bio.lmu.de;martinperniske.
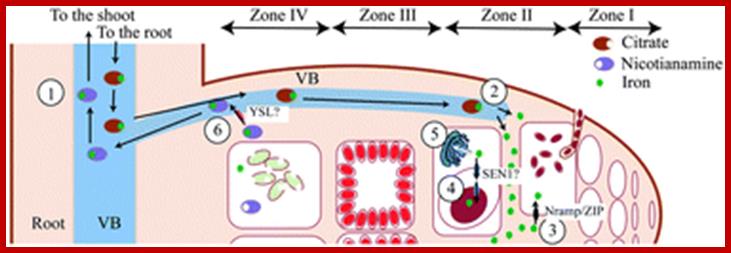
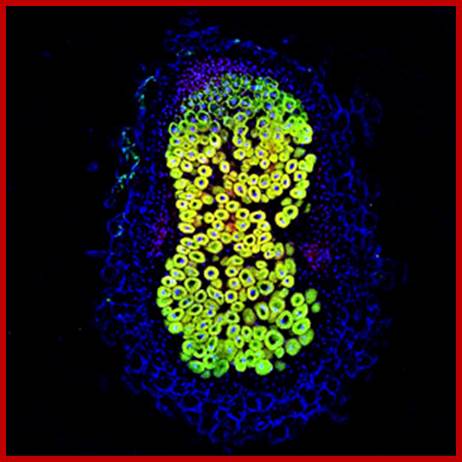
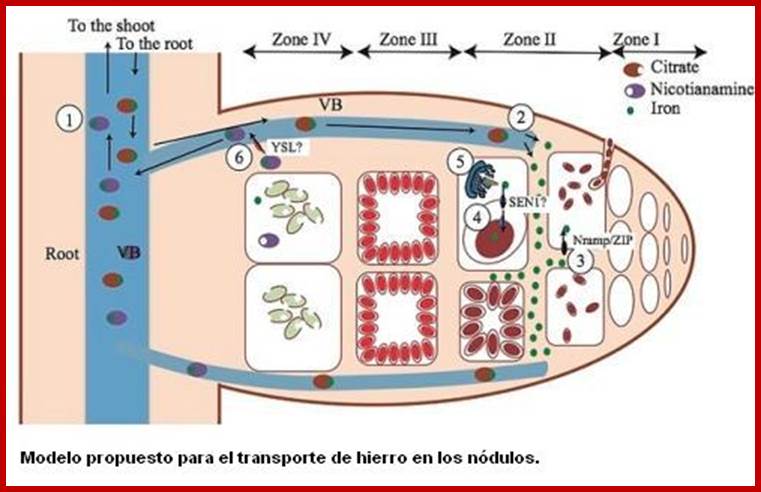
Paramount to symbiotic nitrogen fixation (SNF) is the synthesis of a number of metalloenzymes that use iron as a critical component of their catalytical core. Since this process is carried out by endosymbiotic rhizobia living in legume root nodules, the mechanisms involved in iron delivery to the rhizobia-containing cells are critical for SNF. In order to gain insight into iron transport to the nodule, we have used synchrotron-based X-ray fluorescence to determine the spatio-temporal distribution of this metal in nodules of the legume Medicago truncatula with hitherto unattained sensitivity and resolution. The data support a model in which iron is released from the vasculature into the apoplast of the infection/differentiation zone of the nodule (zone II). The infected cell subsequently takes up this apoplastic iron and delivers it to the symbiosome and the secretory system to synthesize ferroproteins. Upon senescence, iron is relocated to the vasculature to be reused by the shoot. These observations highlight the important role of yet to be discovered metal transporters in iron compartmentalization in the nodule and in the recovery of an essential and scarce nutrient for flowering and seed production. Benjamín Rodríguez-Haas; http://pubs.rsc.org/
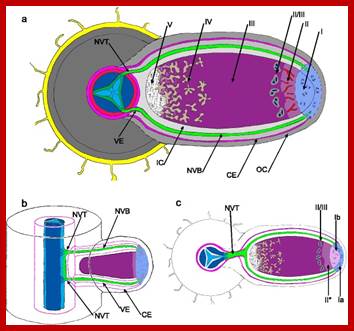
Infection:
Once a particular specific rhizobial strain binds to the host root hair cell, the bacteria induce the formation of infection thread.
http://nichecreator.com/
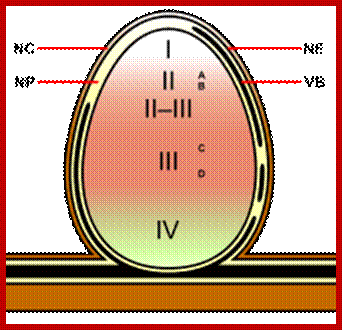
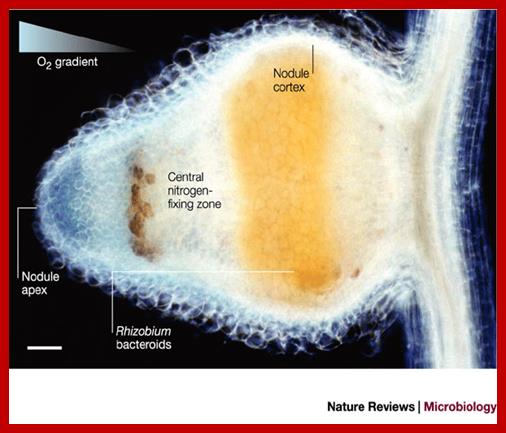
Zone I—the active meristem. This is where new nodule tissue is formed which will later differentiate into the other zones of the nodule.
Zone II—the infection zone. This zone is permeated with infection threads full of bacteria. The plant cells are larger than in the previous zone and cell division is halted.
Interzone II–III—Here the bacteria have entered the plant cells, which contain amyloplasts. They elongate and begin terminally differentiating into symbiotic, nitrogen-fixing bacteroids.
Zone III—the nitrogen fixation zone. Each cell in this zone contains a large, central vacuole and the cytoplasm is filled with fully differentiated bacteroids which are actively fixing nitrogen. The plant provides these cells with leg hemoglobin, resulting in a distinct pink color.
Zone IV—the senescent zone. Here plant cells and their bacteroids contents are being degraded. The breakdown of the heme component of leg hemoglobin results in a visible greening at the base of the nodule. This is the most widely studied type of nodule, but the details are quite different in nodules of peanut and relatives and some other important crops such as lupins where the nodule is formed following direct infection of rhizobia through the epidermis and where infection threads are never formed. Nodules grow around the root, forming a collar-like structure. In these nodules and in the peanut type the central infected tissue is uniform, lacking the uninfected ells seen in nodules of soybean and many indeterminate types such as peas and clovers.
Diagram illustrating the different zones of an indeterminate root nodule ; https://en.wikipedia.org/wiki/Root_nodule
Zone 1; active meristem; Zone II- Infection Zone; Zone III-N2 fixation zone and ZoneIV-Senescent zone; http://nichecreator.com/
http://www.scienceopen.com
Bacterial cells can also enter into the hair cells at the injured surfaces. The infection thread develops from the inner primary cell wall, which grows inwards in the form of invagination enclosing bacterial cells. The infection thread further grows inwards and invades the cortex and finally it finds its way into pericyclic region where the end of the infection thread bursts open releasing bacterial cells. As the infection grows inwards bacterial cells multiply by cell division and the process of multiplication continues even after they are released into the host cells. Bacterial cells assume various shapes and also they aggregate into groups. Such bacterial clusters surrounded by a thin membrane are called bacteroids.
www.nature.com
NODULE FORMATION
With the entry of rhizobial cells into pericyclic cells, if the host cell is a tetraploid cell, the cell undergoes transformation into actively dividing cells otherwise they do not respond to bacterial infection. However, the infected tetraploid cells then divide and redivide to produce a mass of cells which assume nodular form. The growth and the development of a nodule requires the secretion of indole acetic acid (IAA) by the bacterial cells.
INTERACTION BETWEEN BACTERIA AND HOST CELLS
With entry of bacterial cells into host cells, bacterial cellular components stimulate host genome, where globin genes and other related genes get expressed. As a result globin proteins and other factors are synthesized in significant quantities. The globin protein produced in leguminous root nodules is called leghemoglobin, whose amino acid sequence and structure is similar to that of animal globin proteins.
First bacterial nodulation (nod) genes are activated in response to plant secreted signal factors such as flavonoids, resulting in synthesis of nodulation factors by rhizobium. Next ‘nod’ factors elicit nodulation on host roots.
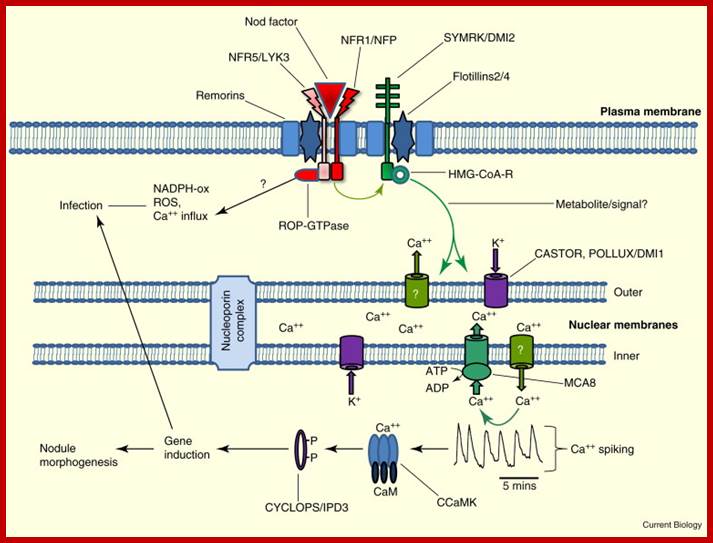
Interaction between rhizobia and plant as host; www.glycoforum.gr.jp; www.cell.com
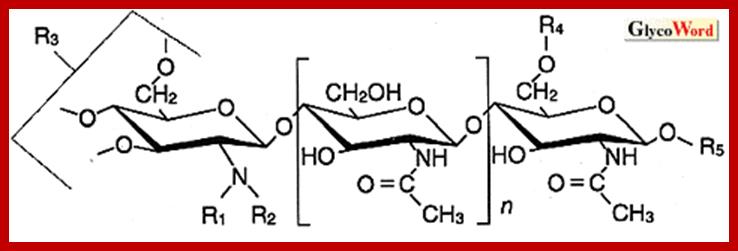
Legume nodulation; Nodule factors; http://www.glycoforum.gr.jp/
On the contrary, host cellular factors inturn activate the expression of nitrogen fixing genes found in rhizobial cells. The ‘nif’ genes remain unexpressed if the rhizobial cells are free from host cells. Though the bacterial cells are associated with the host cells, the genes remain unexpressed if the nitrogen sources like nitrate and ammonia are present in the medium. Only in the absence of them, the N2 fixing genes are expressed. Hence the nitrogenase and other related enzymes are considered as inducible enzymes. Thus, the interaction between the host cellular components and bacterial cellular components is very important in the expression of each other’s genomes for N2 fixation.
NIF GENES AND NITROGENASE
Nitrogen fixing (Nif) genes are a family of 17 genes. The primary enzyme encoded by the nif genes is the nitrogenase complex which is in charge of converting atmospheric nitrogen (N2) to other nitrogen forms such as ammonia which the organism can use for various purposes. Besides the nitrogenase enzyme, the nif genes also encode a number of regulatory proteins involved in nitrogen fixation. The nif genes are found in both free-living nitrogen-fixing bacteria and in symbiotic bacteria associated with various plants. They are located in the bacterial chromosomes but scattered over a length. Among them 10-11 genes are mainly responsible for the synthesis of functional enzyme complex which is made up of larger subunits, smaller subunits and cofactors. Rest of the genes code for the other factors, some of which induce leghemoglobin gene expression in the host cells and the rest are involved in nitrogen fixing activity. The expression of the nif genes is induced as a response to low concentrations of fixed nitrogen and oxygen concentrations (the low oxygen concentrations are actively maintained in the root environment of host plants). The first Rhizobium genes for nitrogen fixation (nif) and for nodulation (nod) were cloned in the early 1980s by Gary Ruvkunand Sharon R. Long in Frederick M. Ausubel's laboratory.

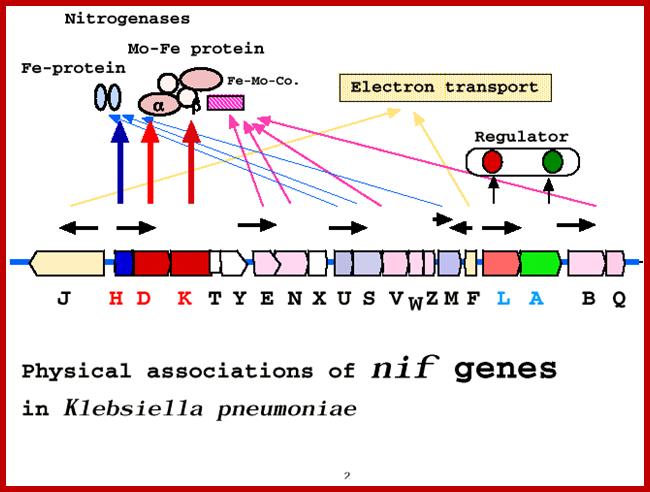
Nif genes map 24 KBp in Klebsiella pneumonias and their roles; http://www.asahi-net.or.jp/
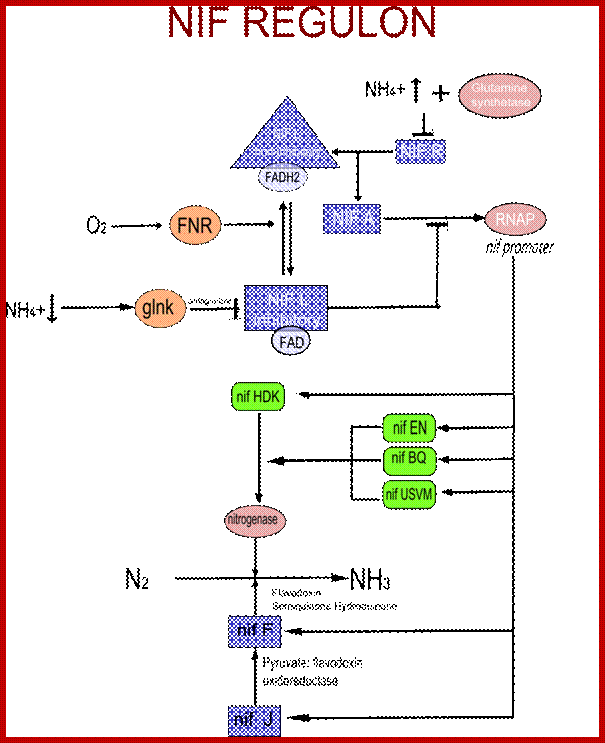
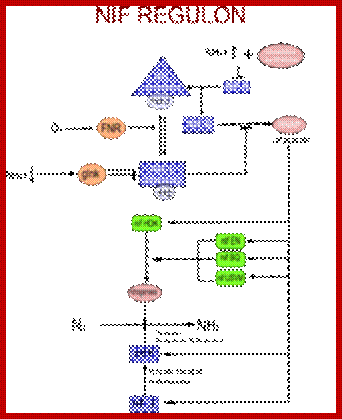
Nif. regulon is the regulatory system to control nitrogen fixation in K. pneumoniae; https://en.wikipedia.org
In most bacteria, regulation of nif genes transcription is activated by the nitrogen sensitive NifA protein. When there isn't enough fixed nitrogen available for the organism's use, NtrC triggers NifA expression, and NifA activates the rest of the nif genes. If there is a sufficient amount of reduced nitrogen or oxygen is present, another protein is activated: NifL. NifL inhibits NifA activity resulting in the inhibition of nitrogenase formation. NifL is regulated by the products of glnD and glnK. The nif genes can be found on bacterial chromosomes, but in symbiotic bacteria they are often found on plasmids or symbiosis islands with other genes related to nitrogen fixation (such as the nod genes).
Nif Regulon: A regulon consists of several operons and each operon might be made up of 2 to many structural genes; here Nif regulon consists of nifRLAA operon, nifHDK operon, nif EN and nifBQ operon, nifJ operon, nifUSVM operon and nif WF operon
The mol. Wt. of nitrogenase enzyme is 250-320 kd. It consists of complex II with 4 large subunits and two small subunits. The small subunit is in association of iron and Mo2 ions as activators. The enzyme is highly sensitive to oxygen and it will be irreversibly destroyed in the presence of oxygen. For its stability and activity, the enzyme has to be maintained in an anaerobic environment within the cell itself. In root nodules, however, leghemoglobin proteins have a dual role to play. While leghemoglobin mop up all the oxygen present in the environs of nitrogenase enzyme enclosed in a membranous vesicle called microsomes, it also provides oxygen for other cellular structures for oxidative mechanisms. In the case of obligate anaerobic bacteria which live in free state, how cells maintain anaerobic condition intracellular is not known and the same is true with the root nodules.
NITROGENASE IS ALSO AN INDUCIBLE ENZYME
As mentioned earlier, nitrogenase enzyme is also an inducible enzyme. In the presence of nitrogen sources like NO3, nitrites and ammonia from cellular environment, this enzyme is expressed through gene activation. In blue green algae, like Nostoc and anabaena, in the absence of above said nitrogen sources, the vegetative cells get transformed into large hyaline cells, whose polar ends will be plugged by some cell wall materials to create an anaerobic environment within the cells.
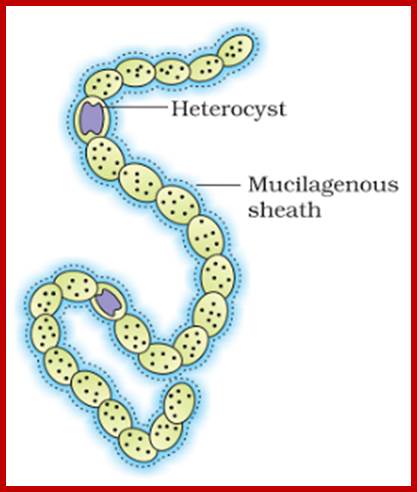
www.knowledgeclass.blogspot.com
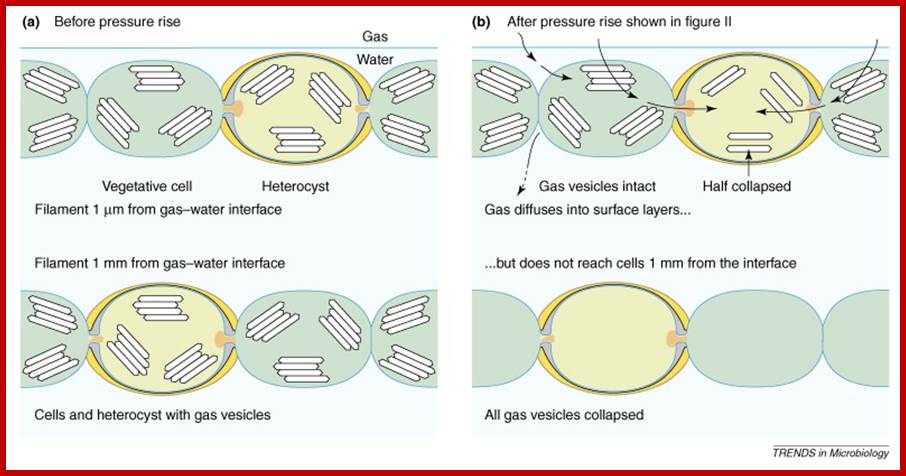
heterocyst cells; Terminal pores for gas exchange; www.cell.com

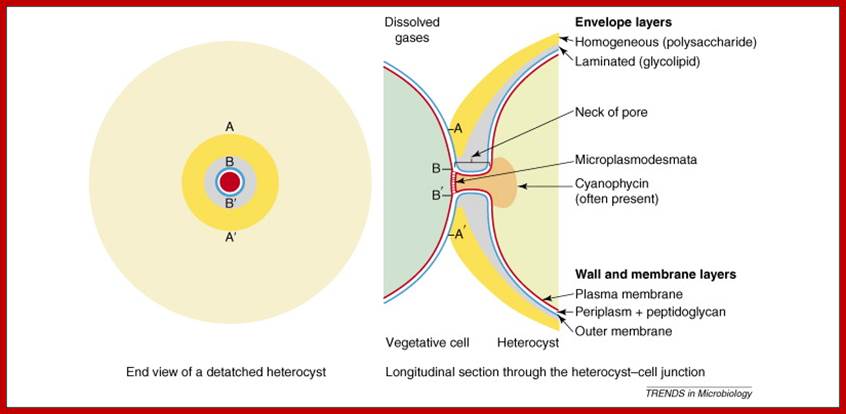
Terminal pores for exchange of gases; www.cell.com
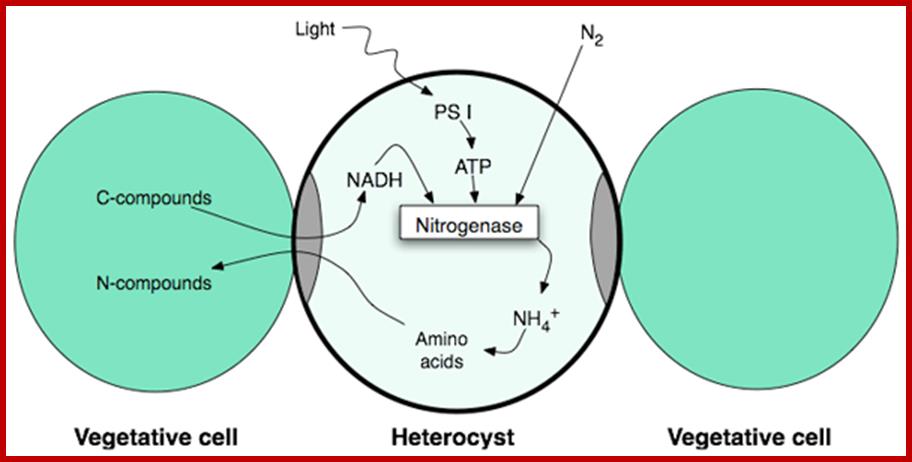
Interaction between vegetative and Heterocyst cells;www.gopixpic.com
MECHANISM OF NITROGEN REDUCTION TO NH3
In order to explain the mechanism of reduction of inert molecular nitrogen to utilizable form i.e. NH4 various theories have been proposed in the past. However, recent studies support the view that the molecular N2 is reduced on the surface of the enzyme nitrogenase in a multistep process. The intermediate products remain bound to the enzyme surface and only the final product ie NH4 is released from the enzyme.

www.meritnation.com; ctayloreco.weebly.com
The reductive power i.e. NADH2 (or NADPH+H) and ATP energy required for this process is supplied by the products of glycolytic pathway or HMP pathway. To begin with, the large subunit part of the nitrogenase enzyme is activated by ATP.
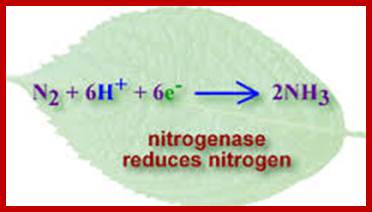
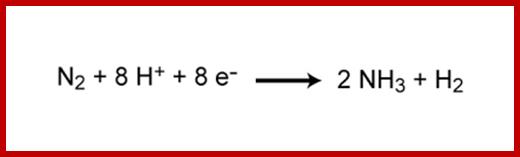
Chemical reaction of Nitrogen fixation; Nature Education; http://www.nature.com/

Nitrogen fixation involves the cleavage of the triple bond of N2 by the enzyme nitrogenase. The ability to fix nitrogen depends on presence of nitrogenase. The only confirmed nitrogen fixers are bacteria (Eubacteria and Archaebacteria).
Nitrogenase activity decreases sharply upon exposure to oxygen and methods have been devised for the protection of nitrogenase. These are physical, behavioral or biochemical such as high respiration rate, scavenging of oxygen, production of leghaemoglobin.
Hetero 8mer A4B2C2; Nitrogenase; www.rcsb.org
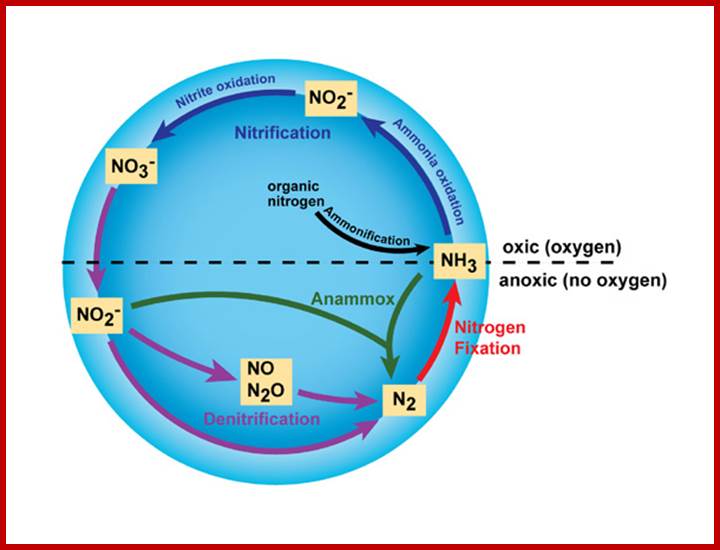
Major transformation in Nitrogen cycle; http://www.nature.com/
Breaking the N2 triple bond: insights into the nitrogenase mechanism; Brett M. Barney et al.
www.id.wikipedia.org
www.pubs.rsc.org
|
N2 + 8H+ + 8e- + 16 ATP = 2NH3 + H2 + 16ADP + 16 Pi |
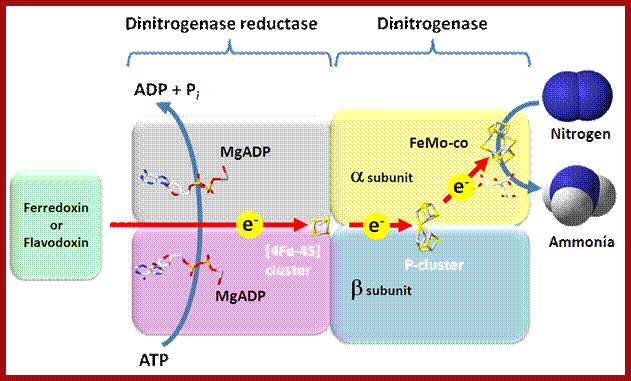
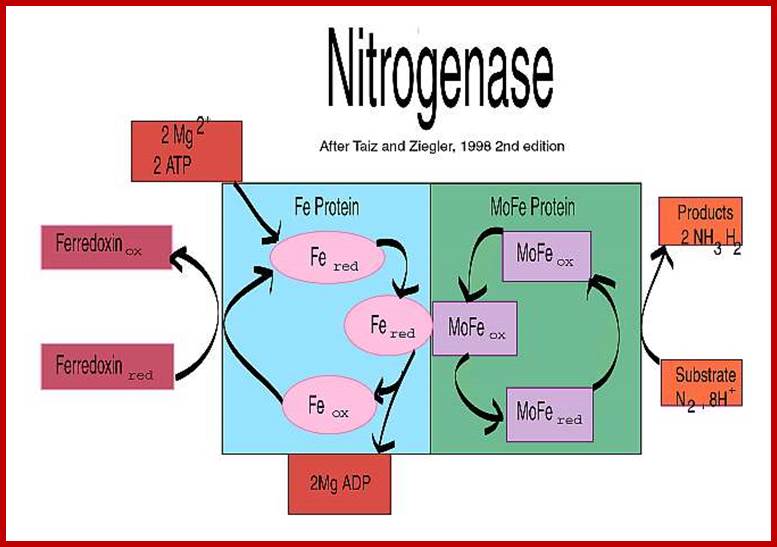
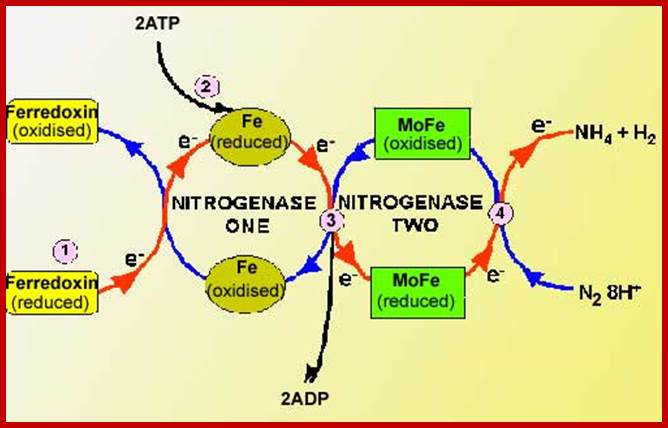
This reaction is performed exclusively by prokaryotes (the bacteria and related organisms), using an enzyme complex termed nitrogenase. This enzyme consists of two proteins - an iron protein and a molybdenum-iron protein (see picture).
http://www.atolls-polynesie.ird.fr/(http://opbs.okstate.edu/~Blair/Bioch4113/
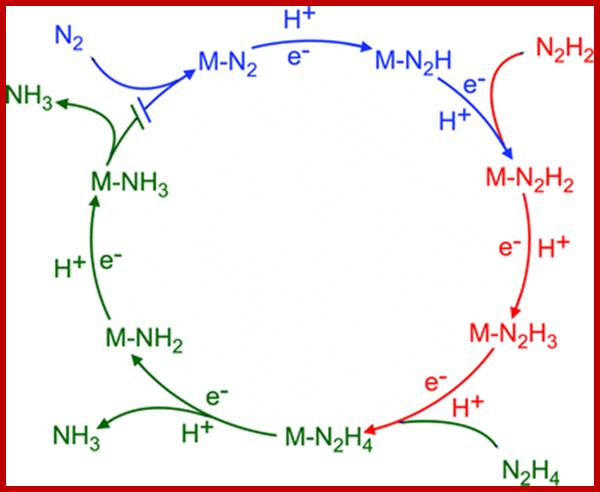
Nitrogenase is the metalloenzyme that performs biological nitrogen fixation by catalyzing the reduction of N2 to ammonia. Understanding how the nitrogenase active site metal cofactor (FeMo-cofactor) catalyzes the cleavage of the N2 triple bond has been the focus of intense study for more than 50 years. Goals have included the determination of where and how substrates interact with the FeMo-cofactor, and the nature of reaction intermediates along the reduction pathway. Progress has included the trapping of intermediates formed during turnover of non-physiological substrates (e.g., alkynes, CS2) providing insights into how these molecules interact with the nitrogenase FeMo-cofactor active site. More recently, substrate-derived species have been trapped at high concentrations during the reduction of N2, a diazene, and hydrazine, providing the first insights into binding modes and possible mechanisms for N2 reduction. A comparison of the current state of knowledge of the trapped species arising from non-physiological substrates and nitrogenous substrates is beginning to reveal some of the intricacies of how nitrogenase breaks the N2 triple bond. Brett M. Barney et al.
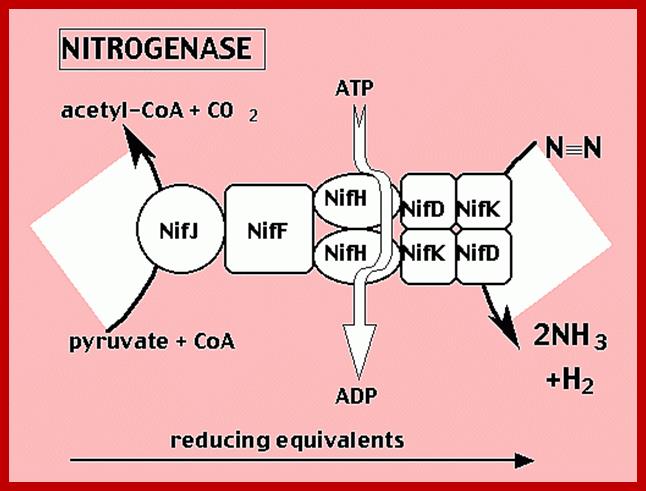
Nitrogenase enzyme complex fixed molcularN2 into Ammonia
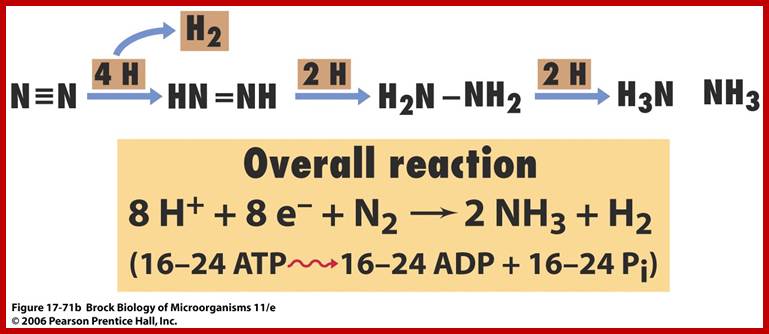
The activated enzyme now accepts molecular N2 and the same binds to the enzymatic surface at specific sites. The energized enzyme now loosens the triple bonds between N atoms, probably through conformational change in the protein structure. Thus, the inert triple bonded N2 is rendered active N=N ready for reductive step. At this juncture, the large complex part of nitrogenase enzyme containing Fe2+ and Mo2+ is reduced by NADH+H. In this oxidation reduction step, the electrons are conveyed to the activated N=N, through Fe/Mo2+. At the same time the activated N=N gets reduced to HN-NH called diimide by another reduction reaction.
With another round of activation of enzyme by ATP, and reduction of HN-NH, is further reduced to Hydrazine i.e. H2N=NH2. In the final round of activation and reduction, the hydrazine gets reduced to 2NH3 which are immediately freed from the surface of the enzyme. So, the reduction of one mole of N2 to two moles of NH3 requires 10-12 moles of ATP, and 3 moles of NADH+H+.
The NH3 thus produced within the bacterial cell is assimilated into glutamate, which is then released into host cells. This way, leguminous plants fix molecular nitrogen to utilizable form of nitrogen.
GENETIC ENGINEERING OF NIF GENES
Nitrogenase genes have been identified and isolated by recombinant DNA techniques from various N2 fixing bacterial cells. Transfer of such genes into higher plant cells is a formidable task. Inspite of it, biologists have succeeded in incorporating such genes into protoplasts of higher plants by incubating protoplasts in a medium containing exogenously supplied nif genes through plasmids, but what is disconcerting in these experiments is that the incorporated genes do not express in the plant cells. Probably the expression of incorporated nif genes required many regulatory gene products and other factors that maintain intracellular anaerobic atmosphere. People are making attempts to make non leguminous plants like paddy, sorghum, wheat, corn, etc., compatible for rhizobial infection to their root system. In some labs, plant genetic engineers are making attempts to hybridize heterocyst cells of Nostoc with higher plant cells. The success of these experiments, if it happens, brings about another super green revolution in the field of agriculture. We have to wait and see for that D-day to be dawn.
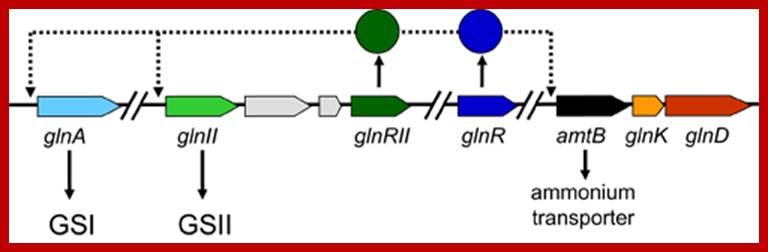
Nif Genes; http://anglerz.com/
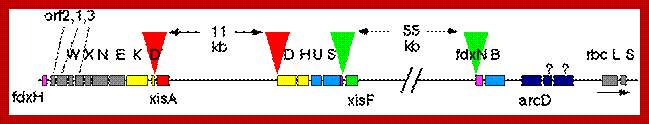
Nif Gene Clusters: http://anglerz.com/
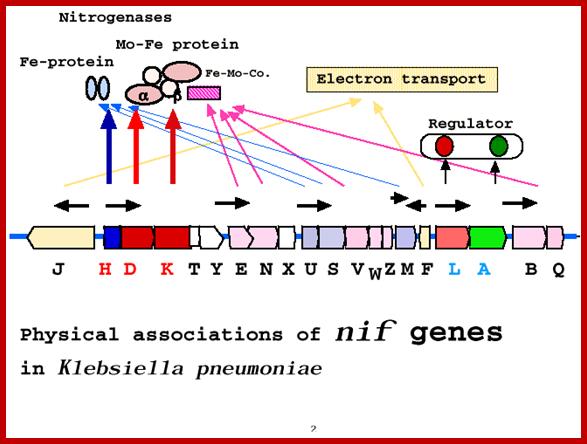
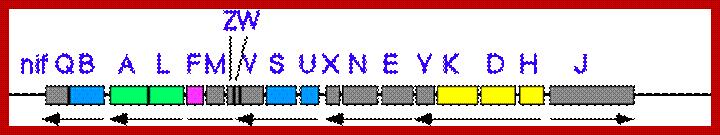
Nif gene map in klebsiella pneumonieae; http://www.asahi-net.or.jp/
Klebsiella nif gene clusters:
In Klebsiella nif fixation is regulated by a nif regulon, which consists of seven operon and 17 genes. The nif regulon consists of seven operons: nifRLA, nifJ, nifHDK,nifEN, nifUSVM, nifWF, nifBQ (Wikipedia).
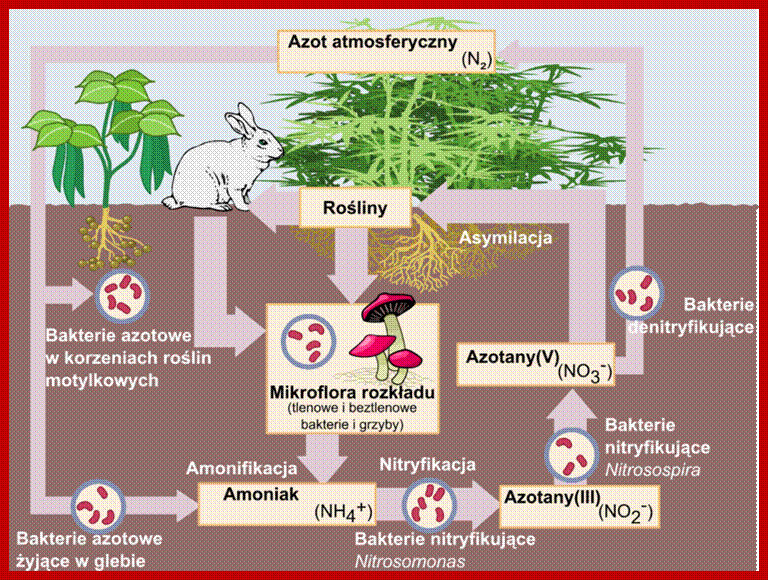
NITROGEN CYCLE
As shown in the self explanatory figures, plants, animals and soil micro organisms bring about the interconversions of inert N2 to utilizable form of N2 and back to inert N2 by various ways and means.
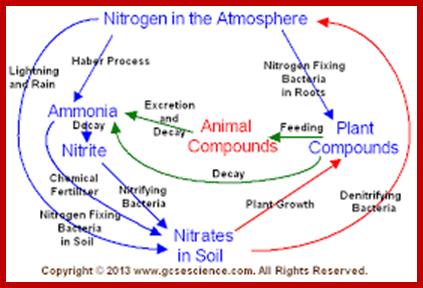
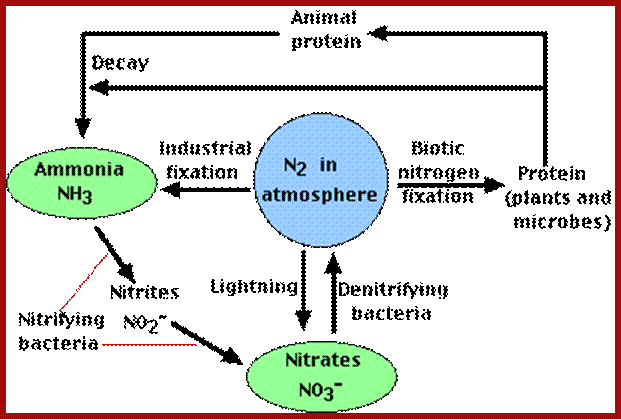
users.rcn.com
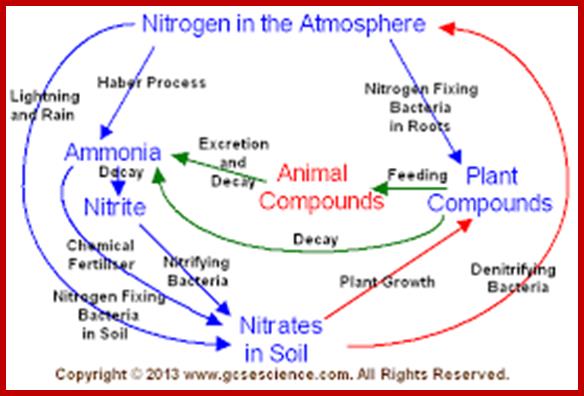
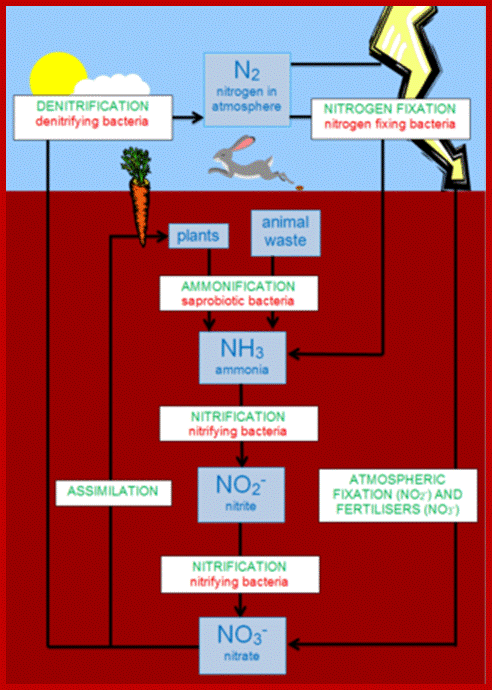
Blue boxed show stores of Nitrogen; green writing for process that occur to move N2 fromone position to the other and red all bacteria involved; https://en.wikipedia.org/wiki/Nitrogen_cycle
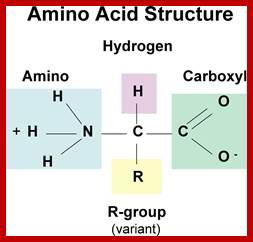
AMINOACID METABOLISM
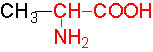
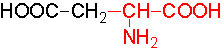
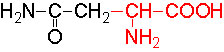
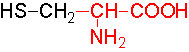
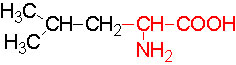
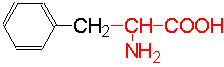
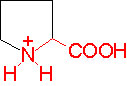
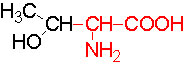
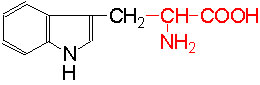
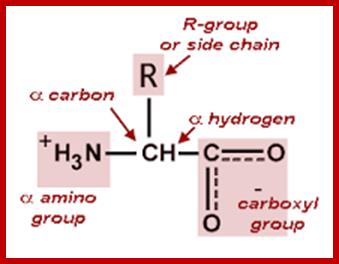
Organic acids containing an amino group at one end and a carboxyl group at the other and are called amino acids. They are one of the most important of cellular components, because they are used in the synthesis of proteins, nitrogenous bases (for nucleic acids), alkaloids, phenolic compounds, porphyrin compounds, flavinoids, pigments, etc. Thus, amino acids play a central role in cellular structures and cellular metabolism. More than 150 amino acids have been identified from various plant sources, but only 20 of them are involved in the formation of proteins, others have different functions.
www.memrise.com; biologie.univ-mrs.fr
PROPERTIES
Amino acids when extracted appear as amorphous powder. They are sparingly soluble in water. With the exception of glycine all others show an asymmetric carbon atom to which one amino group, one carboxyl group, one R-group and one hydrogen are linked. So they exhibit the properties of chirality and isomerism. They also show optical property like D and L forms. Almost every amino acid found in plant or animal proteins have been identified as L amino acids. It is really paradoxical to observe that living organisms have chosen L forms of amino acids for cellular metabolism but with regard to carbohydrates they have selected D forms as carbohydrate units. It is difficult to explain why and how life forms have selected L-form of amino acids and D forms of carbohydrates. Interestingly the D forms of amino acids are used in the production of some important cyclic or linear proteins, some of which are antibiotics.
Amino acids, because of their ionizable property show different electrical charges in the same molecule. Under different pH conditions, they can exist either as basic ions, acidic ions or neutral ions. The neutral ions are also called amphoteric or Zwitter ions. In fact, the pH at which an amino acid exists as a Zwitter ion is referred to as isoelectric point. Different amino acids show different isoelectric points and they have to be determined by titration against known concentration of a base or an acid. Amphoteric amino acids are also called ampholytes and they are of greater use in chemical industry and medical research.
Further more, the R groups present vary from amino acid to amino acid, because they may contain additional basic amino groups, carboxyl groups, sulfhydril groups, hydroxyl groups, aromatic groups or CH3 groups. Depending upon the nature of R groups different amino acids have been identified.
DETECTION OF AMINOACIDS
Almost all amino acids react with Ninhydrin, a coloring reagent, and produce purple coloration. Proline ad hydroxyproline produce yellow color action with Ninhydrin reagent. Using Ninhydrin reaction methods, it is possible to identify individual amino acids but also, they can be estimated quantitatively. Mixtures of amino acids can be separated by paper chromatography or by automatic amino acid analyzers and they can be identified as well as estimated. Another interesting method that is employed for identification of specific amino acids is by the use of 1’ fluoro 2.4 dinitro benzene which under mild alkaline conditions react with amino acids to produce 2,4 dinitrophenyl derivatives. In finger printing of a polypeptide chain this method is very useful.
CLASSIFICATI8ON OF AMINOACIDS
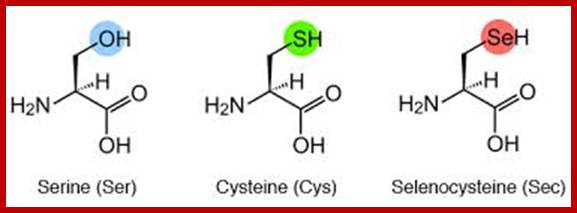
Almost every amino acid found in cellular proteins has been identified as L form. The total number of such amino acids found in all biological system is just twenty. But one more is added, i.e. Selenocysteine.
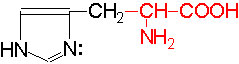
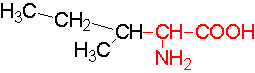
The said twenty-one different amino acids can be easily identified because of different R groups. Based on this L form of amino acids has been classified into the following types and each one of them shows characteristic features; aliphatic, aromatic, heterocyclic, hydroxyl, acidic, basic and sulphur containing.
The said twenty different amino acids can be easily identified because of different R groups. Based on this L form of amino acids has been classified into the following types and each one of them show characteristics features.
|
Amino Acid structure |
Name |
Symbol |
|
|
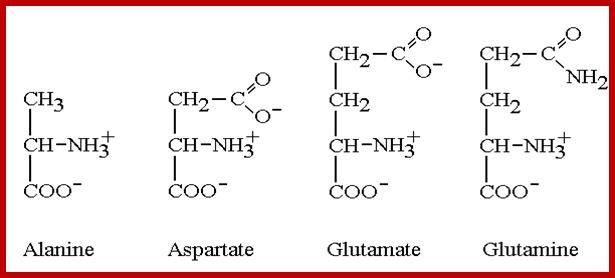
Alanine |
A |
|
|
Aspartic |
D |
|
|
Aspargine |
N |
|
|
Cysteine |
C |
|
|
|
|
|
|
Glutamic |
E |
|
|
Glutamine |
Q |
|
|
Glycine |
G |
|
|
Histidine |
H |
|
|
Isoleucine |
I |
|
|
Leucine |
L |
|
|
Lysine |
K |
|
|
Methionine |
M |
|
|
Phenylalanine |
F |
|
|
Proline |
P |
|
|
Serine |
S |
|
|
Threonine |
T |
|
|
Tryptophan |
W |
|
|
Tyrosine |
Y |
|
|
Valine |
V |
|
|
Selenocyteine |
Sc |
21st amino acid- Seleno cysteine;
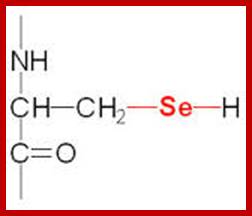
www.themedicalbiochemistrypage.org
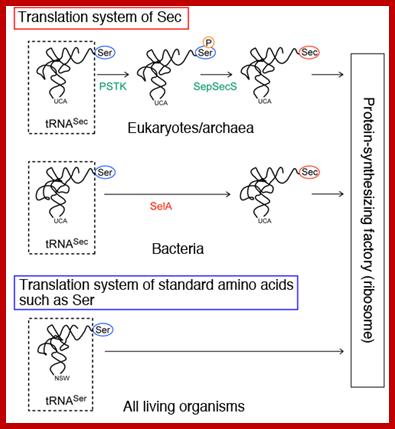
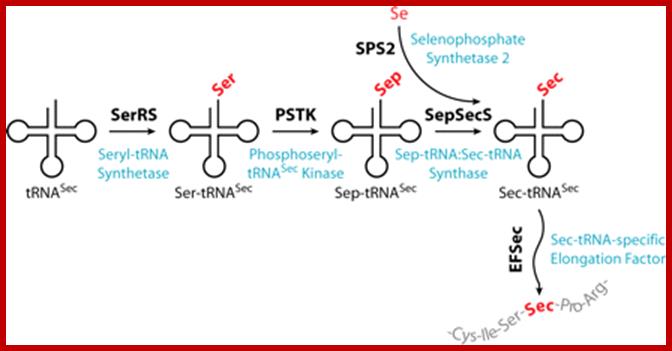
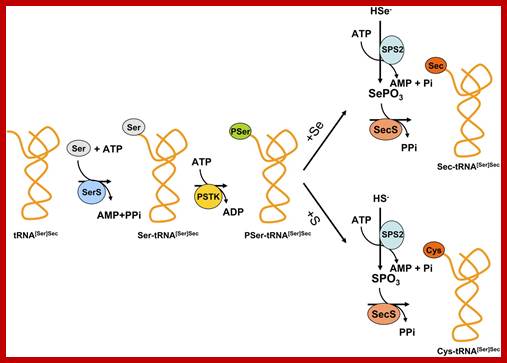
It is synthesized when serine is added to its tRNA, then serine is converted to cysteine and then cysteine into Seleno cysteine. This amino acid is required to insert selenium into specific proteins Selenocysteine synthesis requires Sel-A enzyme. Selenocysteine incorporation in eukaryotic proteins occurs cotranslationally at UGA codons (normally stop codons) via the interactions of a number of specialized proteins and protein complexes. In addition, there are specific secondary structures in the 3′ untranslated regions of selenoprotein mRNAs, termed SECIS elements that are required for selenocysteine insertion into the elongating protein. One of the complexes required for this important modification is comprised of a selenocysteinyl tRNA [(Sec)-tRNA(Ser)Sec] and its specific elongation factor identified as selenoprotein translation factor B (SelB). SelB is also commonly called eukaryotic elongation factor, selenocysteine-tRNA-specific (EEFsec or EFsec). The protein that is involved in the interaction of the SECIS element with the (Sec)-tRNA(Ser)Sec if referred to as SECIS binding protein, SBP2. Additional proteins involved in synthesis pathway include two selenophosphate synthetases, SPS1 and SPS2, ribosomal protein L30, and two factors that have been shown to bind (Sec)-tRNA(Ser)Sec identified as soluble liver antigen/liver protein (SLA/LP) and SECp43.
Selenocysteine biosynthesis and incorporation; The first steps involve the activation of serine onto the (Sec)-tRNA followed by enzymatic conversion to selenocysteine generating (Sec)-tRNA(Ser)Sec. Next the (Sec)-tRNA(Ser)Sec is bound by SelB and the complex is incorporated into the translational machinery aided by SBP2 (not shown). The elongating protein is transferred to the selenocysteinyl-tRNA via the action of peptidyl transferase as for any other incoming amino acid and normal elongation continues. www.themedicalbiochemistrypage.org
Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii; Oselov SV1, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN.
Known eukaryotic selenocysteine (Sec)-containing proteins are animal proteins, whereas selenoproteins have not been found in yeast and plants. Surprisingly, we detected selenoproteins in a member of the plant kingdom, Chlamydomonas reinhardtii, and directly identified two of them as phospholipid hydro peroxide glutathione peroxidase and selenoprotein W homologs. Moreover, a selenocysteyl-tRNA was isolated that recognized specifically the Sec codon UGA. Subsequent gene cloning and bioinformatics analyses identified eight additional selenoproteins, including methionine-S-sulfoxide reductase, a selenoprotein specific to Chlamydomonas: Chlamydomonas selenoprotein genes contained selenocysteine insertion sequence (SECIS) elements that were similar, but not identical, to those of animals. These SECIS elements could direct selenoprotein synthesis in mammalian cells, indicating a common origin of plant and animal Sec insertion systems. We found that selenium is required for optimal growth of Chlamydomonas: Finally, evolutionary analyses suggested that selenoproteins present in Chlamydomonas and animals evolved early, and were independently lost in land plants, yeast and some animals.
http://www.spring8.or.jp/
Selenocysteine insertion; http://www.pnas.org/
http://bioinformatica.upf.edu/
Functions and benefits Selenocysteine:
§ The important functions of selenocysteine in proteins are its anti –oxidant activity. This is due to its lower pKa and higher reduction potential.
§ It is also used in the preparation of variety of vitamins and lots of other supplements.
§ It is also fortified with livestock feeds.
§ Our body utilizes selenocysteine to form selenium, which is believe to play important role in preventing mercury toxicity as well as enhance liver functions.
§ Selenocysteine is not directly incorporated into other proteins. It provides it function on its own. For this reason, it is highly reactive and not used in the same way as the body uses other amino acids.
§ In some cases, a marked decrease in catalytic activity of an enzyme is observed when a selenocysteine residue is replaced with cysteine. This substitution caused complete loss of glycine reductase selenoprotein A activity.
§ Selenocysteine is the 21st naturally occurring amino acid, and is coded for by the RNA codon UGA, which is normally a “stop” signal, but is modified in some organisms to create selenocysteine by a subsequent RNA loop, which is interpreted by a group of genes called the sel group, which are activated by the loop in the mRNA and produce molecule of tRNA for selenocysteine.
§ Selenocysteine is an organic selenium compound found naturally in some plants such as garlic, onions, broccoli and wild leeks grown in high selenium soil.
§ Seleno cysteine is also found in wheat, oats, corn, rice, and soybeans
Selenocysteine deficiency;
- People deficient with selenium have lean body mass, prone to premature aging, weaken the immune system, making the body more susceptible to illness, as well as lead to heart disease or hypothyroidism and often cancer. https://blissreturned.wordpress.com
Biosynthesis of amino acids
Plants are capable of synthesizing amino acids in every living cell but most of the primary amino acids are synthesized in roots and leaves. The ammonia produced by the reductive steps of NO2, NO3, or N2 are toxic if they are accumulated in the cells. Hence, the ammonia is immediately used up in the synthesis of amino acids. If there is any excess of NH3 it is stored in amides, from which the same can be recovered.
The most important pathway by which amino acids are synthesized is reductive amination leading to the synthesis of glutamate. The other pathways like transamination and carbonyl phosphate reactions are called secondary pathways.
REDUCTIVE AMINATION PATHWAY
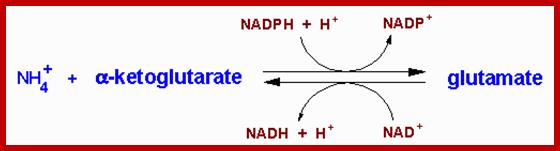
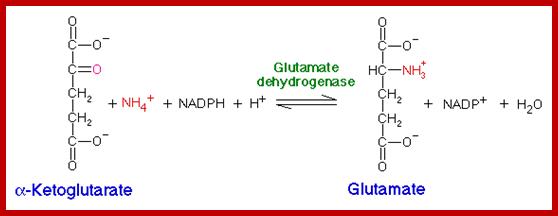
The most important carbon compound that acts as an acceptor of amino group is ketogluterate which is an intermediate product of Kreb’s cycle. Though alfa ketogluterate is mainly used in Kreb’s cycle to generate energy, depending upon the need of the cells or tissues, it is also drawn into reductive amination pathway for the synthesis of glutamate. This pathway is catalyzed by an enzyme known as ketogluterate dehydrogenase which brings about amination as well as reduction. The mol. Wt. of the enzyme is 320,000 Daltons and it is an allosteric enzyme. That means it is a regulatory enzyme modulated by specific factors. The reducing power used in this reaction is NADH2 in non chlorophyllous tissue) or NADPH2 in chlorophyllous tissue.
www.themedicalbiochemistrypage.org
www.imgarcade.com
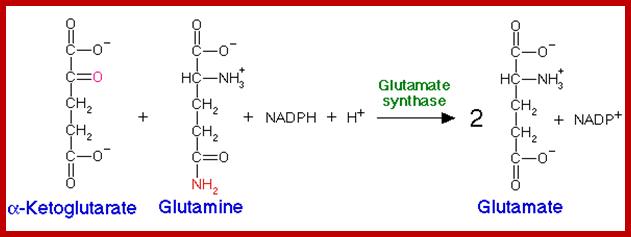
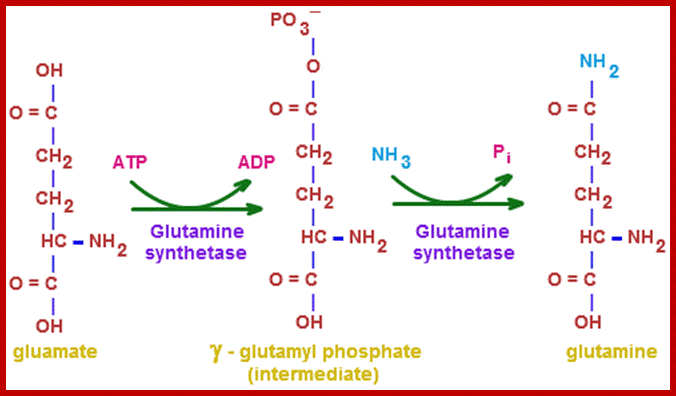
www.themedicalbiochemistrypage.org
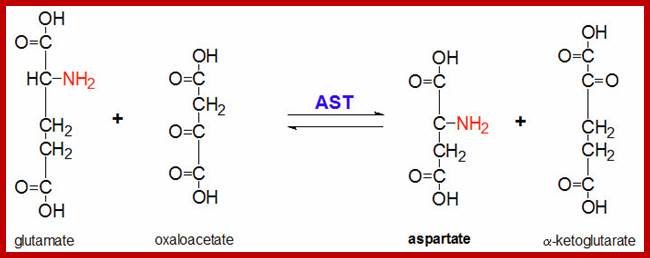
http://chemistry.tutorvista.com/; www.studyblue.com; the medical biochemistry page.org
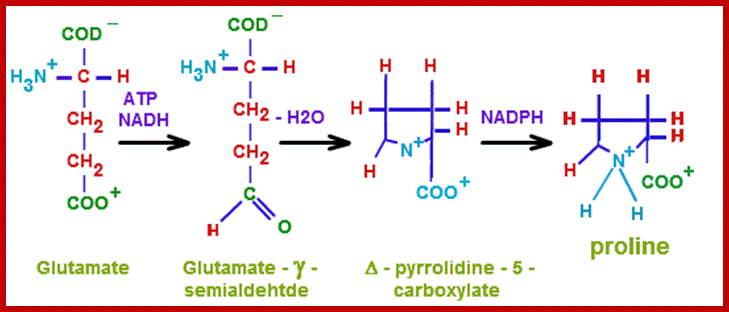
http://chemistry.tutorvista.com/
To begin with alfa ketogluterate, in the presence of NH4 spontaneously reacts and gets converted to alfa immunogluterate. Then alfa imminogluterate is reduced by the dehydrogenase to produce glutamate.
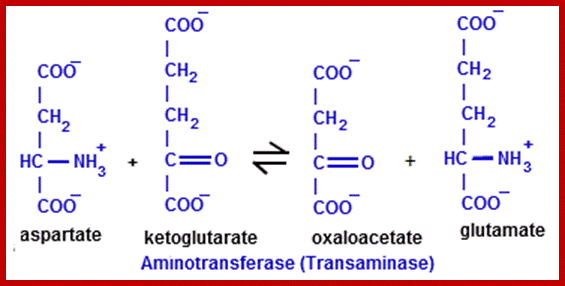
Similarly, the other keto acids like pyruvate and oxoloacetate are also used in reductive amination by specific amino-reductases resulting in the formation of respective amino acids. Pyruvate yields alanine and OAA yields aspartate. But when labeled NH3 is provided to plant tissues, most of the label (90%) ends up in glutamate; only a little quantity is found in alanine and aspartate. Thus, glutamate synthesis acts as the major of pathway in amino acid synthesis. Nonetheless glutamate acts as the donor of amino group for the synthesis of other amino acid.
AMIDE SYNTHESIS:
Plants have some unique mechanism by which excess NH3 is fixed as amides such as glutamine and asparagine. The amide synthesis is performed by specific amide synthetases. The glutamine synthetase is a complex allosteric enzyme regulated by specific factors. This enzyme requires ATP as the energy source for its activity in which extra NH3 group is added onto the additional carboxyl unit present in R group. When ammonia is present in sufficient quantities, most of the labeled ammonia ends up in glutamine and glutamic acid.
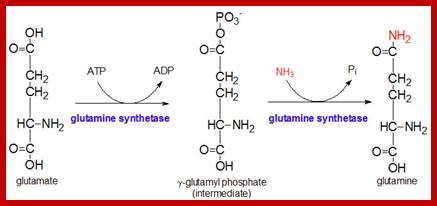
themedicalbiochemistrypage.org6
nitrogenes.cropsci.illinois.edu
www.imgarcade.com
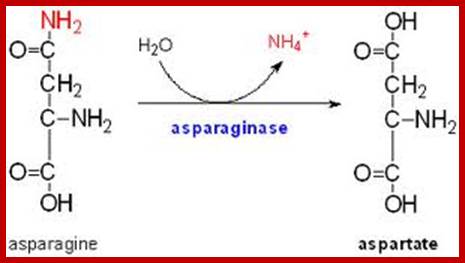
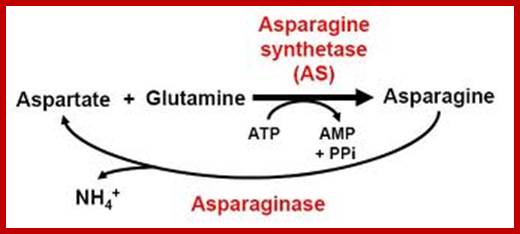
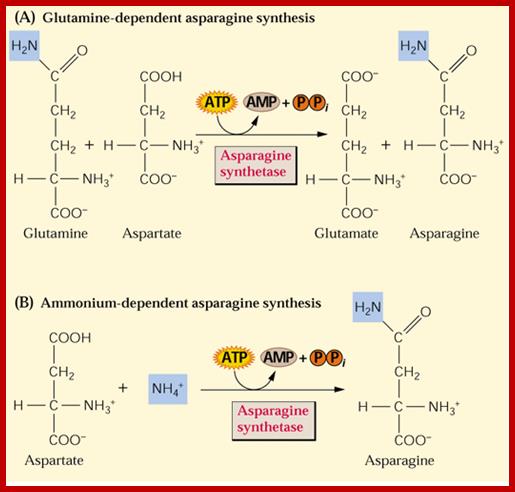
Similarly, Aspargine is also synthesized by amination reaction, which also requires activation energy supplied by ATP. The enzyme involved in this reaction is Aspargine synthetase, which is also an allosteric enzyme.
The amides thus synthesized act as reserve components. Whenever there is a need for NH3 for the synthesis of amino acids either by reductive amination or by other processes, glutamine and Aspargine are subjected to deamination reactions by the activity glutamine or Aspargine deaminase enzymes and the NH3 released is used. It is important to note that synthesis and glutamine hydrolysis is not reversible reactions because the enzymes involved are different.
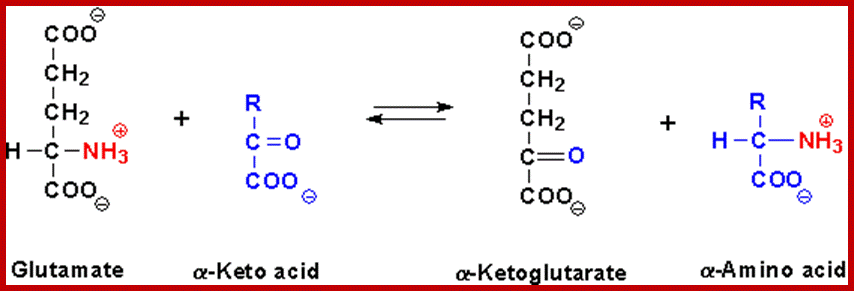
TRANSAMINATION PATHWAY;
The transaminase enzymes are capable of transferring amino group from the donor amino acid to the acceptor ketonic organic acids. The coenzyme involved in this reaction is pyridoxal phosphate, which has a unique role in picking only amino group and then transferring to the ketonic group of the acceptor molecule.
In plants, of the activities of aspartate amino transferase and alanine amino transferase have been studied. Similarly, the synthesis of serine in peroxisomes found in C3 plants is also brought about by transaminase activities.
SYNTHESIS OF CARBOMOYL PHOSPHATE AND ARGININE
Plants have another unique pathway where they utilize respiratory CO2 and free NH3 to synthesize carbomoyl phosphate. The enzyme responsible for this process is carbomoyl phosphate synthetase. It is a mitochondrial enzyme and its activities has been studied in the leaves of phaseolus, pea and castor plants. The mechanism and properties of this enzyme is similar to that of animal mitochondrial carbomoyl phosphate synthetase. This enzyme requires ATP for its activity.
Carbamoyl phosphate is a very important compound; it is used in the synthesis of ornithine, citruline and Arginine. It is also used in the synthesis of nitrogenous bases.
Plants have been known to utilize urea as the source of NH3 for amino acid syntheses. But recent experiments, on chlorella pyrenoids and chlorella ellipsoides, have clearly demonstrated that urea is directly used in a condensation reaction with ornithine to produce arginine. However, the other properties of this enzyme have not been fully characterized.
Urea + ornithine -> Arginine
INTERCONVERSIONS:
Plants possess various metabolic pathways in which starting from simple amino acids and other organic acids, they are capable of synthesizing all amino acids required for protein synthesis and other metabolic processes. Similarly, amino acids are also used in gluconeogenesis produce some intermediary compounds or to produce more energy. However these pathways are very well regulated.
Most of the pathways that lead to the synthesis of various amino acids other than glutamic acid, aspartate, alanine, are multistep reactions. For example, glutamic acid is used to synthesize amino acids like proline, arginine, tryptophan or histidine in multistep reactions. Similarly, aspartate is metabolized to produce threonine or lysine; pyruvate is used in the synthesis of valine; phosphoenol pyruvate is converted to phenylalanine or tyrosine and anthralinic acid is metabolized to produce tryptophan, etc. Most of these pathways are regulated either at the enzymatic level or at the level of gene expression. For example, the synthesis of proline starting from glutamate exhibits feedback inhibition. Similarly, the pathways leading to the synthesis of threonine and lysine starting from aspartate are inhibited by their and product.
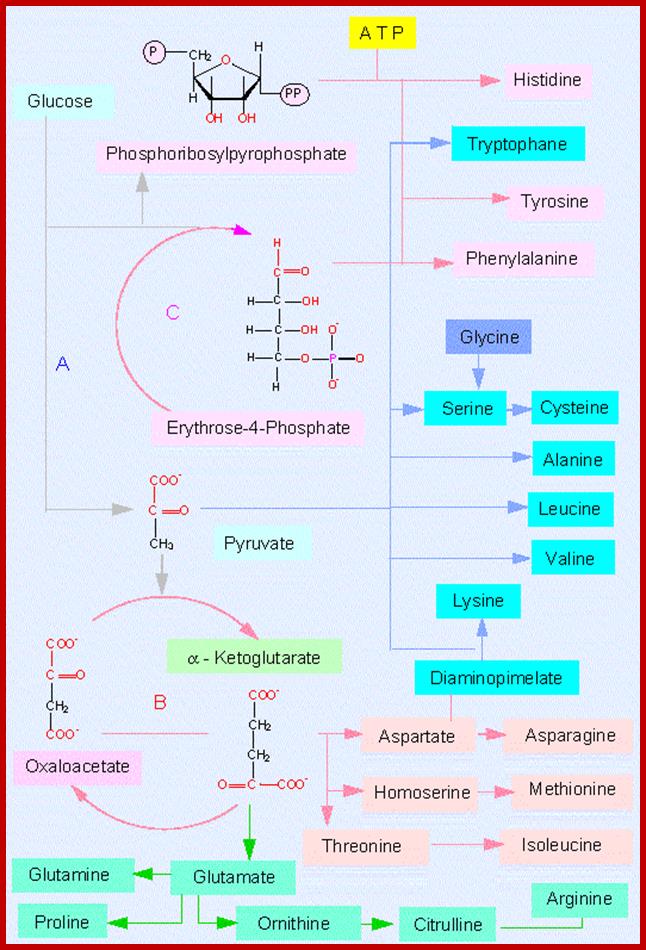
Glucose to amino acids
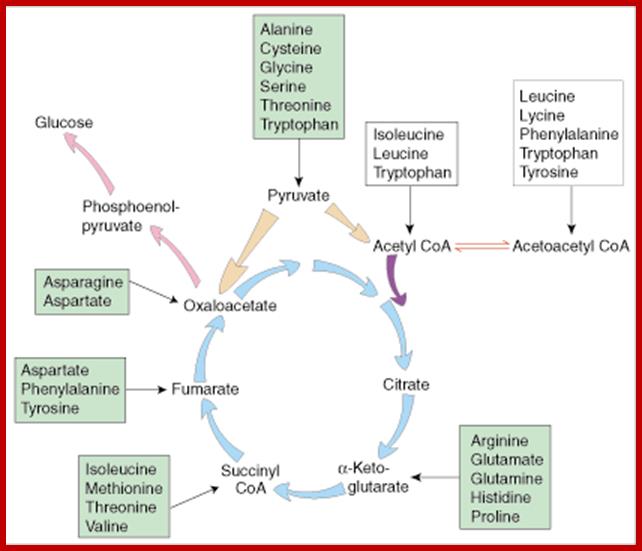
The intermediates can then be converted to oxaloacetate, the main precursor for gluconeogenesis. The following amino acids are glucogenic: alanine, cysteine, glycine, serine, threonine, tryptophan, asparagine, aspartate, phenylalanine, tyrosine, isoleucine, methionine, threonine, valine, arginine, glutamate, glutamine, histidine, and proline. http://www.sparknotes.com/
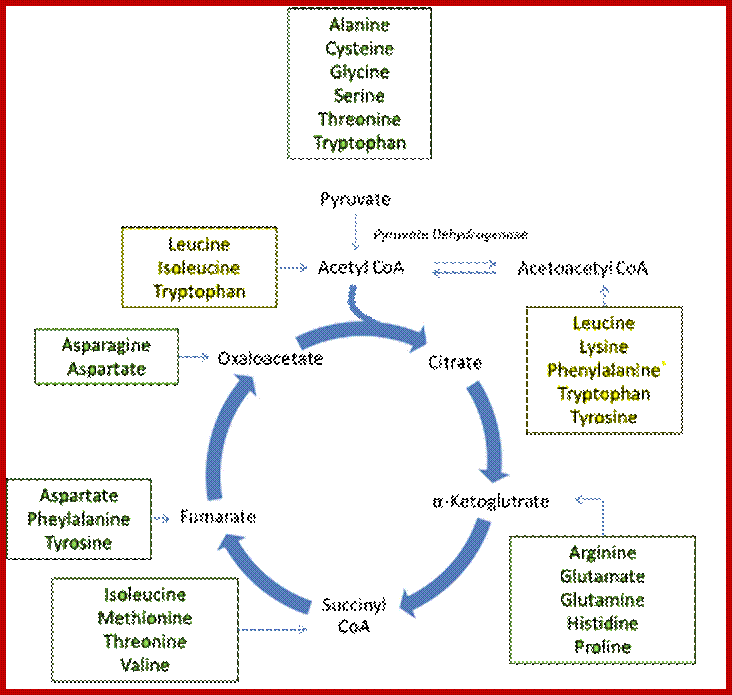 . Glucogenic amino acids are listed in GREEN
boxes and ketogenic amino acids are listed in YELLOW boxes.
. Glucogenic amino acids are listed in GREEN
boxes and ketogenic amino acids are listed in YELLOW boxes.
; https://www.khanacademy.org
Synthesis of histidine is not only regulated at the enzyme level but also it is controlled at the gene level. In the absence of histidine in the culture medium, entire sequence of genes responsible for all the enzymes that are required for histidine synthesis are expressed and the pathway operates leading to the production of histidine. Once histidine accumulates in sufficient quantities it binds to the first enzyme of the pathway and inhibits its activity. This is called as feedback inhibition. At the same time, histidine also binds to the inactive apo-repressor and makes it active. Then the active repressor binds to histidine operator gene, thereby the whole sequence of genes responsible for histidine synthesis are repressed (Refer regulation of gene expression is prokaryotes).
Such feed back inhibition of multistep pathways is also found in threonine to isoleucine and tryptophan pathways. With regard to tryptophan as an end product not only acts as an inhibitor at the enzyme level but also acts as the co-repressor in its gene expression.
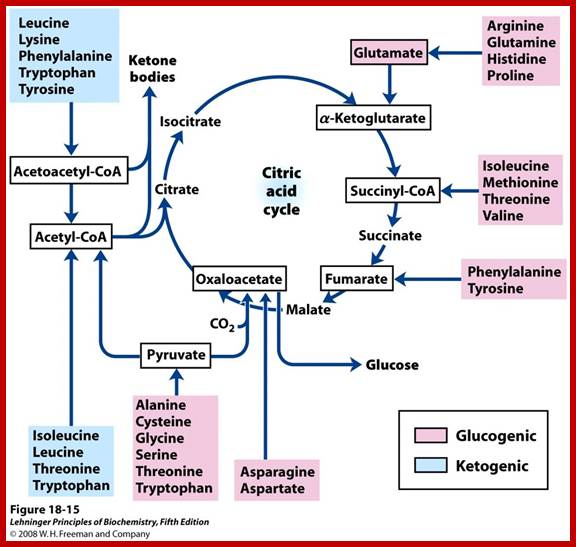
On the other hand, depending upon the demand or the intracellular conditions, many amino acids found within the cells are interconverted or metabolized to various organic compounds and some of them are virtually drawn into citrate cycle or they may be used in glycogenic pathways. As shown in the figure one can find how different groups of amino acids are drawn into Kreb’s cycle. Thus, interconversions also help in producing many key intermediary products required for specific metabolic pathway or products. Such pathways are highly regulated.
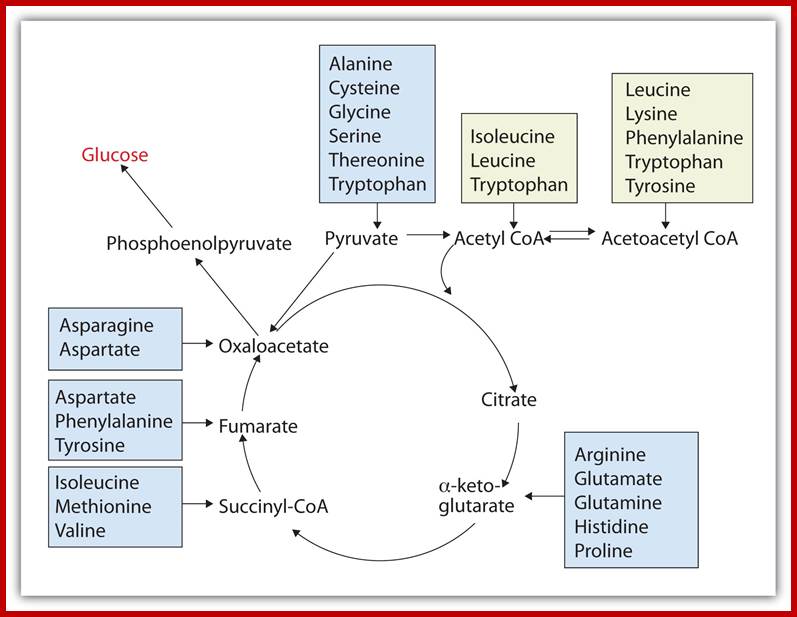
Krebs cycle is the key for synthesis of different amino acids. chemwiki.ucdavis.edu
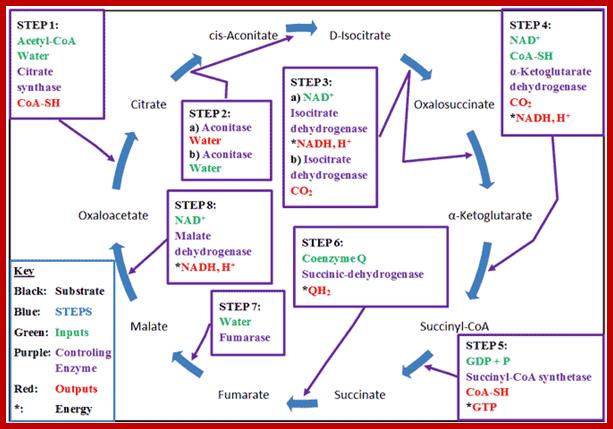
Kreb’s cycle overview; www.wyzant.com
The Krebs Cycle (also known as the Citric Acid or Tricarboxylic Acid (TCA) cycle is the process through which aerobic cellular metabolism occurs. Hans Krebs received the 1953 Nobel Prize in Medicine for his “discovery” of the citric acid cycle. This cycle involves a series of reactions involving a (1) a substrate, Oxaloacetate, that is modified in every reaction, (2) Acetyl–CoA, from which energy is extracted, (3) energy transport reactants, which collect the extracted energy, and (4) the controlling enzymes, which regulate the steps of the cycle. This cycle is ubiquitous in living organisms, single and multi-celled, both plants and animals — including humans. Organizationally, the process is often divided into 8 steps, one for each controlling enzyme, usually beginning with the combination of the Oxaloacetate substrate to the Acetyl–CoA, which is produced from either glycolysis or pyruvate oxidation.
Proteins: Nature’s Nano-Machines; https://amit1b.wordpress.com
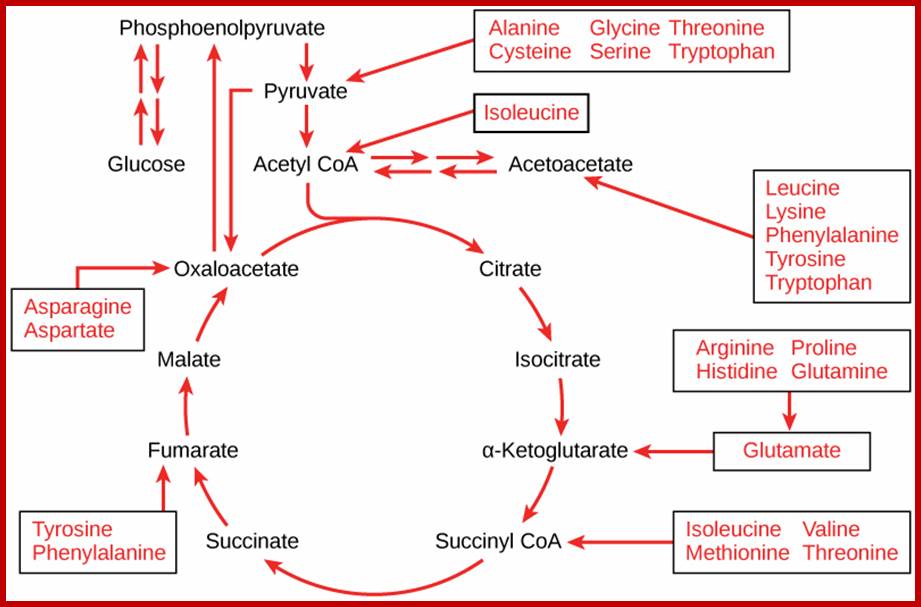
Connections of carbohydrates, proteins and lipid metabolic pathways; http://voer.edu.vn/m/connections-of-carbohydrate-protein-and-lipid-metabolic-pathways/5a
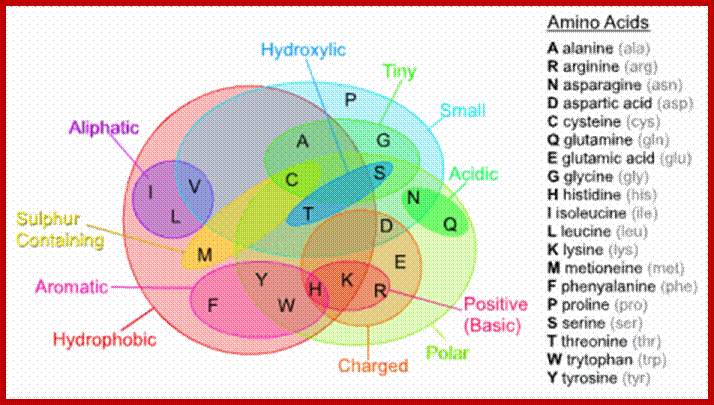
The 20 natural amino acids clustered by their physical-chemical properties (taken from Esquivel et al. (2013)); https://amit1b.wordpress.com