APOPTOSIS 2
Genes and Mechanism:
Death transducing receptors:
Any factor, in any form that impinges on cells has to be first recognized by specific receptors and then the message has to be transduced to internal milieu to asses the signal and if needed execute molecular actions.
Receptors are cellular sentinels, located on the surface of the cell membranes and scan all the external signals that are impinged upon all the time. Thousand of such specific receptors to specific cell type are found located on their cell surface. They span the whole cell surface and they are found in different shapes with different structure and different functions.
Apoptotic receptors are varied and many, such as Fas-R (CD95), TNF-R1 and 2, Apo-1, rpr gene (Reaper in Drosophila), TRAIL1, TRAIL 2 (TNF related apoptosis inducing ligand receptors DR4 and DR5).
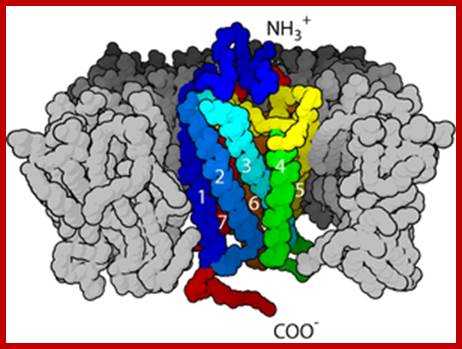
G-protein-coupled receptor- It is seven transmembrane receptor proteins;
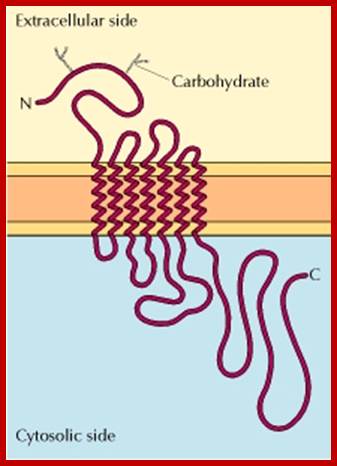
Structure of a G protein-coupled receptor. The G protein-coupled receptors are characterized by seven transmembrane ‘α’ helices. http://www.ncbi.nlm.nih.gov/
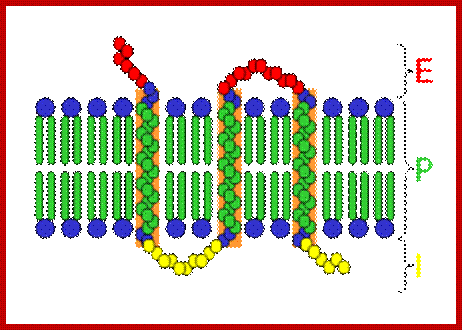
Extra cellular domain, PM- plasma membrane domain and I= Intracellular domain; https://en.wikipedia.org
![]()
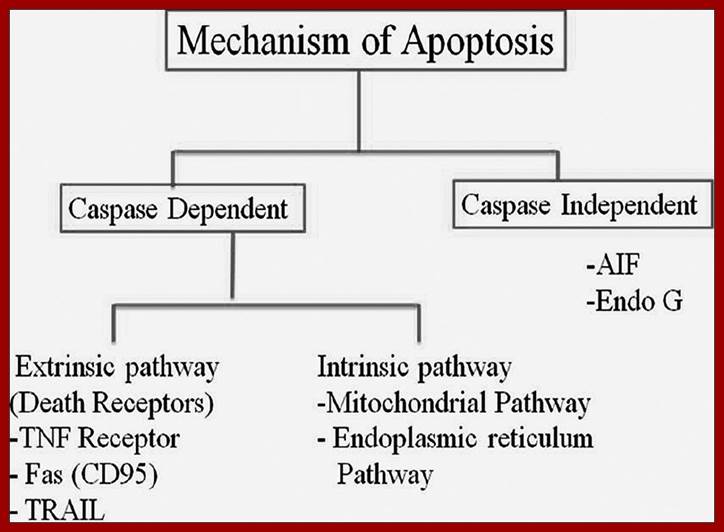
Schematic representation of mechanism of apoptosis; http://www.jofs.in/

http://www.jofs.in/
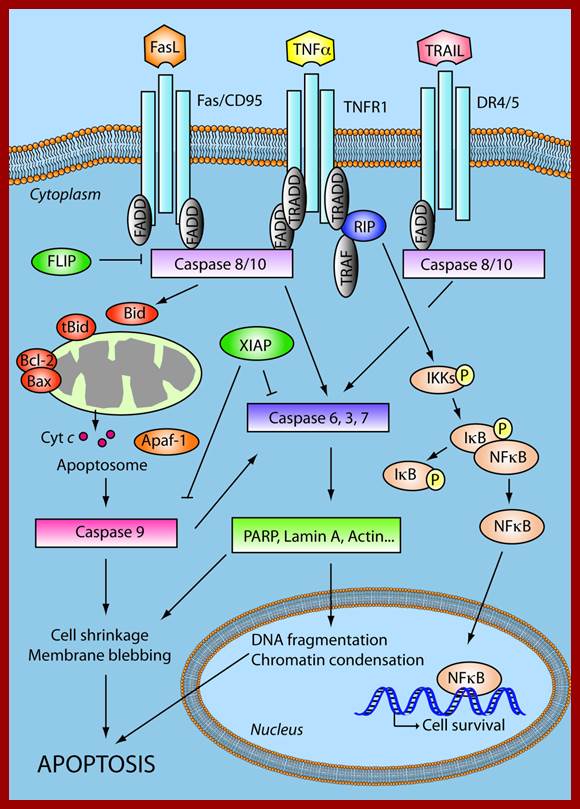
Apoptosis Signalling network: The extrinsic apoptosis pathway is activated upon ligand binding to death receptors (TNFR1, Fas/CD95, DR4/5). This results in activation of a caspase cascade and eventually cleavage of both cytoplasmic and nuclear substrates. TNFR1 may promote survival signaling through activation of NFκB. The intrinsic pathway involves release of apoptotic proteins from the mitochondria, formation of the Apoptosome and subsequently caspase activation. Members of the BCL-2 protein family are involved in regulation of the intrinsic apoptotic pathway. The extrinsic and the intrinsic pathways converge in a caspase cascade that results in cellular shrinkage, DNA fragmentation and eventually apoptosis. These pathways are highly deregulated in GBMs. Tumour necrosis factor receptor (TNFR), Tumour necrosis related apoptosis-inducing ligand (TRAIL), TNFR type 1-associated death domain protein (TRADD), Death receptor (DR), Fas-associated protein with death domain (FADD), TNFR associated factor (TRAF), Receptor interacting protein (RIP), FLICE-like inhibitory protein (FLIP), X-linked inhibitor of apoptosis protein (XIAP), Nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB), Inhibitor of κB (IκB), IκB kinases (IKKs), cytochrome c (Cyt c), Apoptotic protease activating factor 1 (Apaf-1).;http://www.molecular-cancer.com/
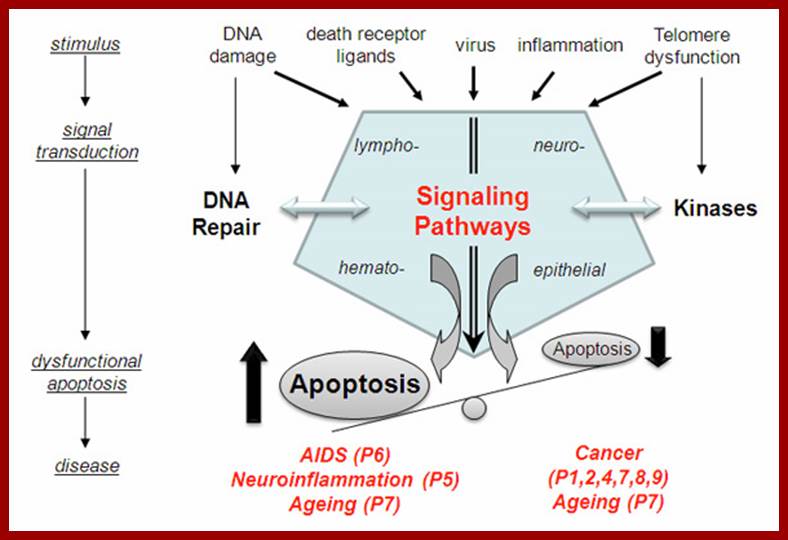
The role of apoptosis signaling pathways in human diseases; www.uniklinik-ulm.de
The receptors are located in plasma membrane with external domain, and a cytosolic domain and a transmembrane domain. For example, Fas-R forms trimers (actually dimer) with the binding of a ligand Fas-L. When a specific signal, binds to a specific receptor on to external domain, leads to certain conformation changes in the receptor proteins. This change is critical and the same is manifested at cytosolic side of the membrane. In the case of Fas-R binding leads to trimerization, it further induces more clustering at cytosolic surface. The cytosolic domain interacts with respective cytoplasmic components and executes actions there off. The cytosolic domain found at carboxy terminal of Fas-R and other related receptors protein contain a 80 amino acid sequence, more or less same (28% conserved) among their related members. This region is called death domain or DD. Interaction between the death domains and cellular components results in activating a set of proteases.
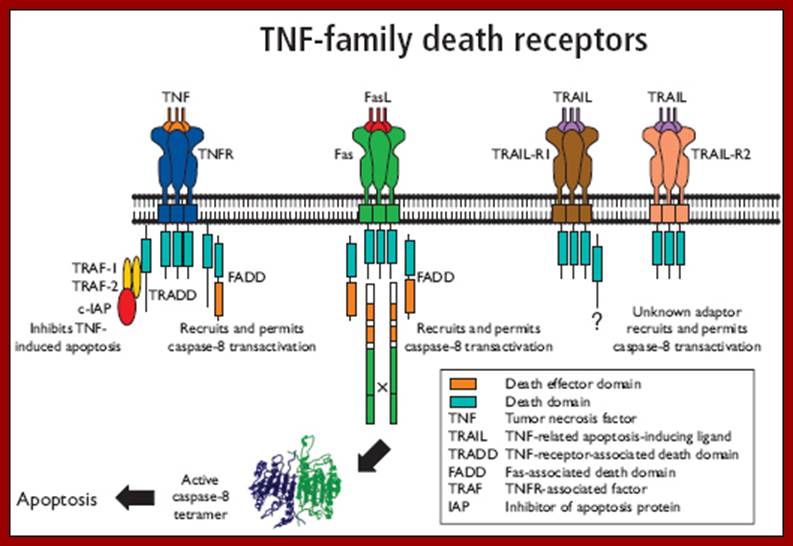
Apoptosis Cell Membrane Receptors - Death Receptors; www.abdserotec.com
www.webbooks.com
This
region is called death domain or DD. Interaction between the death domains and
cellular components results in activating a set of proteases; www.webbooks.com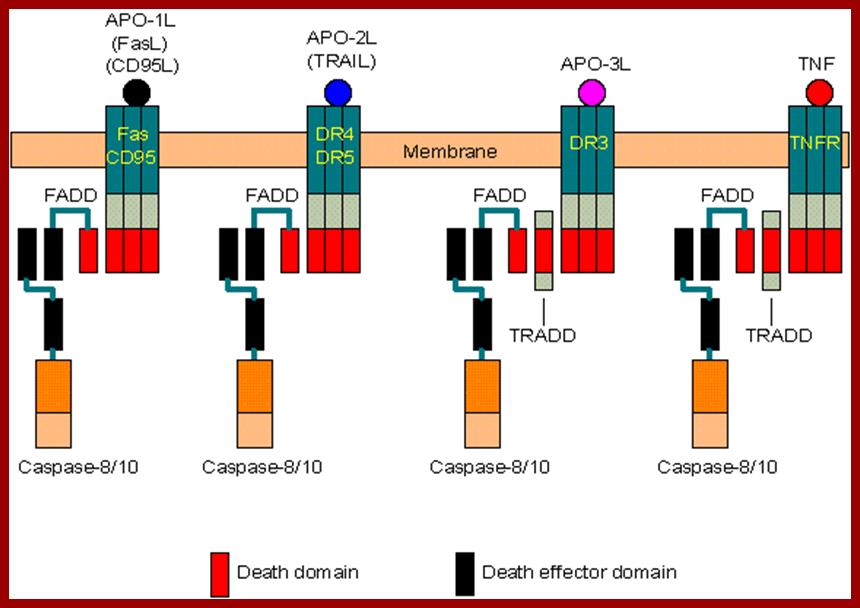
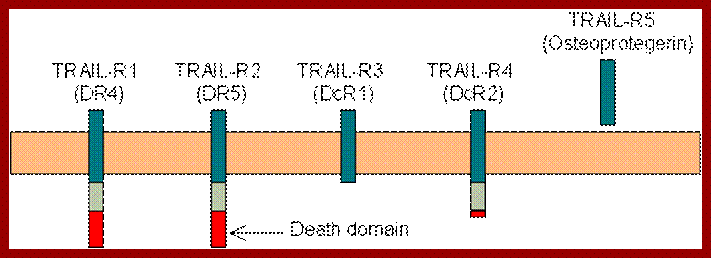
Schematic drawing of TRAIL receptors, which can be divided into two categories: Death receptors: TRAIL-R1 and TRAIL-R2 contain the death domain, capable of inducing apoptosis. Decoy receptors: TRAIL-R3 and TRAIL-R5 lack the death domain while TRAIL-R4 contains a truncated non-functional death domain. These three receptors can bind to TRAIL, but cannot induce apoptosis. TRAIL-R5 is secreted to the extracellular fluid. All other receptors are transmembrane proteins; www.Web books.com
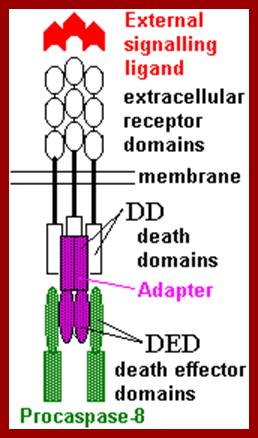
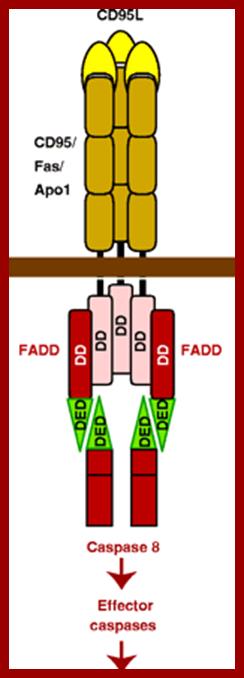
Both the above diagrams represent the signal, docking on to the receptors and interaction of cytosolic death domain with adapters and with Caspase-8, www.webbooks.com
Cell death related genes:
There are a number of genes involved in causing death of cells. Among them proteases play an important role. Genetic studies in C. elegans (Ces), Drosophila and human beings resulted in identifying 14 cell death genes called Ced genes.
· Ces1, Ces2, ecl1 genes determine cell death specificity. Ces-1 causes death of 4 cells in pharynx. Ces 2 causes death of neuronal cells, but mutation in the gene prevents the death of few neurons.
· Egl–egg laying, these cells die in males.
· Proteases, with Cysteine as an active site, that cleaves proteins at carboxyl side of aspartate residue, are called Caspases (Ced). Caspases are a family of 14 or more proteins; Ced 1 to Ced 14. All of them are synthesized as pro-Caspases. Caspases are grouped into two, the first group consists of Caspases related to inflation; such as casp-8, 9 & 10, they have long pro-domains and initiate cell death and the second group consists of casp-3, 6 &7 few more, they have short pro-domains and cleave various structures. They are involved in responding to cell death signal, they are Ced 3 and Ced 6-10.
· Ced 3, Ced4, Ced8 and Ced9 control and execute death.
· Ced4 codes for a calcium binding protein.
· Ced3 is Cysteine protease; it is the most potent protease of all other Ced proteins. This has a homology with that of interleukin 1 b converting enzyme called ICE, which is also a Cysteine protease but required for the death of lymphocytes. Absence of this does not prevent apoptosis. Ced3 recognizes a sequence –YVAD (Tyrosine, Valine, Alanine, and Aspartate) and ICE recognize DEVD sequence (Aspartate, Glutamate, Valine, and Aspartate). Ced9 can protect the cell from death or it can be deadly. This has a homology with Bcl2. Ced 9 is also associated with membranes such as mitochondria, nucleus and ER.
· Ced1, Ced2, ced5, ced6, ced7 and Ced10 control the engulfment of the degraded cellular components by macrophages.
· Nuc1 is an endonuclease, controls degradation of engulfed matter by activated macrophages.
· Transcriptional activators- HLF, DBP and TEF/VBP.
· Transcriptional repressors-E4BP4, Giant, and Ces2.
· Bcl2 an oncogene, it is a member of multi protein complex; it has many related gene members. Bcl2 is a transmembrane protein localized in the outer membrane of mitochondria, nuclear membranes and endoplasmic membranes.
· Bax: it is associated with Bcl2 in membranes and causes the release of Cyt.C.
· Myc and p53, depending upon circumstances they cause survival of cells or induce the cells to death.
· BID, it is an inhibitor protein.
· APAF1, it is an apoptosis protease-activating factor.
· Cytochrome–C, it is mitochondrial protein; with the release of this protein from peri-mitochondrial space, cyt-C activates the whole process of cell death
· FADD: FAS-R associated death domain factor, it is associated with FAS-R. at cytosolic side.
· TRADD: TNFR α1 associated death domain factor, it is associated with TNF a1 receptor at cytosolic side.
|
Name |
KDs |
Target |
Activation |
Domain |
Substrates |
Function/s |
|
Caspase1, ICE |
45920+10 |
WHED |
TPLD, FED |
CARD |
Pro-IL, self |
Against inflammation |
|
Caspase2 |
48(18+12 |
DVAD |
DNKD |
CARD |
Self |
Initiates |
|
Caspase3 |
32(17+12) |
DMQD |
ESMD |
|
PARP, PK, self |
Initiator |
|
Caspase4 |
|
EHD, LEVD |
LEED |
CARD |
Pro-IL, self |
inflamation |
|
Caspase5 |
|
EHD |
WVRD, LEAD |
Self |
|
inflamation |
|
Caspase6 |
34 |
VEHD |
DVVD |
|
|
|
|
Caspase7 |
34(20+12) |
DEVD |
IQAD |
DED |
PARP, ICAD |
Effector |
|
Caspase8 |
53(18+11) |
ETD+ |
VET+ |
DED |
PARP, self |
Initiator |
|
Caspase9 |
50 |
LEHD |
PEPD |
CARD |
PARP, self |
Initiator |
|
Caspase10 |
55(17+12) |
IEHD |
IEHD |
DED |
Pocaspase3, 7 |
Initiates |
|
Caspase11-14 |
|
|
|
|
|
Swelling |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
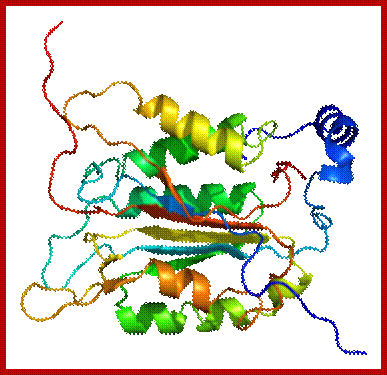
Caspase1; en.wikipedia.org
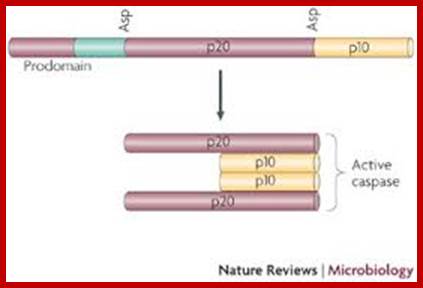
Caspases are a class of cysteine-aspartyl proteases that are synthesized as inactive precursor enzymes or proenzymes (see the figure). These proteases typically lie dormant in the healthy cell and, in response to cell-death stimuli, are converted, either by proteolytic cleavage or by recruitment into large complexes, into active enzymes. Once activated, caspases cleave their substrates after conserved aspartate residues. The effects of caspases on cellular substrates bring about the biochemical and morphological features of apoptosis.
Caspase proenzymes contain three domains: an amino-terminal prodomain; a large subunit that contains the active-site cysteine within a conserved QACXG motif (p20); and a carboxy-terminal small subunit (p10). Two cleavage events at aspartate (Asp) residues are required to activate caspases. The first divides the proenzyme into large and small caspase subunits, and the second removes the N-terminal prodomain. The resulting functional caspase is a tetramer of two large (p20) and two small (p10) subunits.
Caspases are divided into two groups. Initiator caspases (such as caspase 8 and caspase 9) activate the effector caspases and have long prodomains that allow them to interact with death effector domains (DEDs) or caspase recruitment domains (CARDs) in adaptor proteins such as FAS-associated death domain protein (FADD) and apoptotic protease-activating factor 1 (APAF1) (reviewed in Ref. 106). Effector caspases (such as caspase 3) function primarily to cause the morphological features of apoptosis. Specifically, activation of caspase 3 leads to the breakdown of several cytoskeletal proteins, cleavage of poly (ADP-ribose) polymerase (PARP) and degradation of ICAD (inhibitor of caspase-activated DNase), thereby releasing CAD, which cleaves cellular DNA. Activation of caspase 3 and PARP cleavage, by western blot and immunohistochemistry, and DNA fragmentation, by gel electrophoresis and TUNEL, are frequently used as apoptosis assays in vivo. Penny Clarke & Kenneth L. Tyler; www. Nature.com
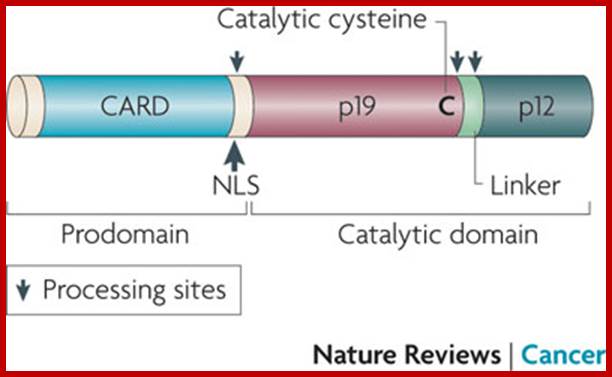
Activation of CASPASE-2; Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more?
Caspase 2 activation occurs during both intrinsic (mitochondrial) and extrinsic (death receptor) cell death signalling. However, it has remained controversial whether caspase 2 is an initiator or effector caspase. Caspase 2 belongs to a group of caspases that contain a caspase activation and recruitment domain (CARD) in their amino-terminal prodomain (see the figure on the right). As CARD is a protein–protein interaction domain, it is found in caspases that are activated by their recruitment to specific caspase activation complexes, such as the 'apoptosome' that recruits and activates caspase 9 and the PIDDosome that recruits and activates caspase 2 (Refs 3,4,41). When caspase 2 is present at a high concentration, its activation can occur by CARD-mediated dimerization or oligomerization, without any need for accessory or adaptor proteins. The initial activation of caspase 2 seems to be mediated by dimerization not processing41. This generates a partially active enzyme that is further activated by auto processing (self-cleavage). Additional processing, which is presumably mediated by active effector caspases (caspases 3 and 7) results in the generation of caspase 2 p19 and p12 subunits. Given that caspase 2 can be processed by caspases 3 and 7, and that loss or inhibition of these caspases or their upstream activators (caspase 9 and APAF1) leads to an inhibition of caspase 2 cleavage (as shown by immunoblotting), caspase 2 is also thought be a downstream caspase. Fully mature caspase 2 forms a dimer in solution, which consists of two p19 and two p12 subunits NLS, nuclear localization;.Sharad kumar; www.nature.com
Targets of Caspases in Cells:
Nucleus: Nuclear lamins, nucleoplasmins, SR proteins, hnRNPs, some Transcription factors such as RNA Pl (pstram) factors, MDM2, RB and inhibitors such as p27 and p21.
DNA: Repair enzymes, including RAD and others, PARP, Topoisomerases, and inhibitors of Caspases (iCAD/DFF45-Caspase activated DNase or DNA fragment factor).
Cytoskeleton structures: Actins, Gelsolins, and pectrins, Keratins.
Cytoplasmic components: beta catenins, Bcl2.
Protein kinases: DNA dependent protein kinase like ATM/ATR, Protein kinase C, CAM kinase, MAPs and ERK kinases, protein kinase-B, Raf1
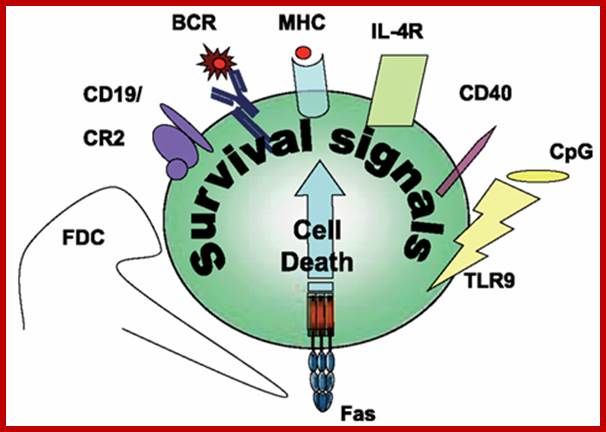
Survival signals rescuing Fas-induced apoptosis in B cells.;dx.doi.org
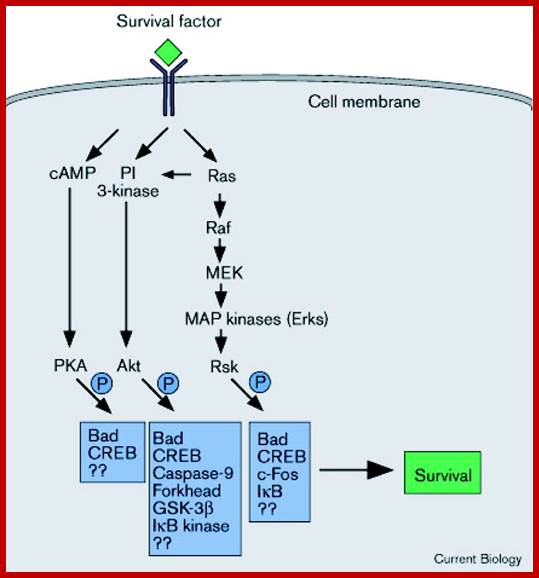
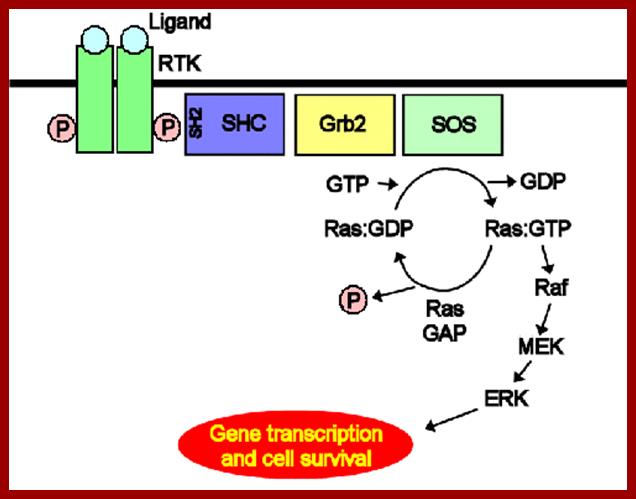
RSK1 mediates MEK-MAP kinase cell survival signal; The diagrams show what happens when cells are signaled by cell surviving signals; Akiko Shimamura et al; www.cell.com
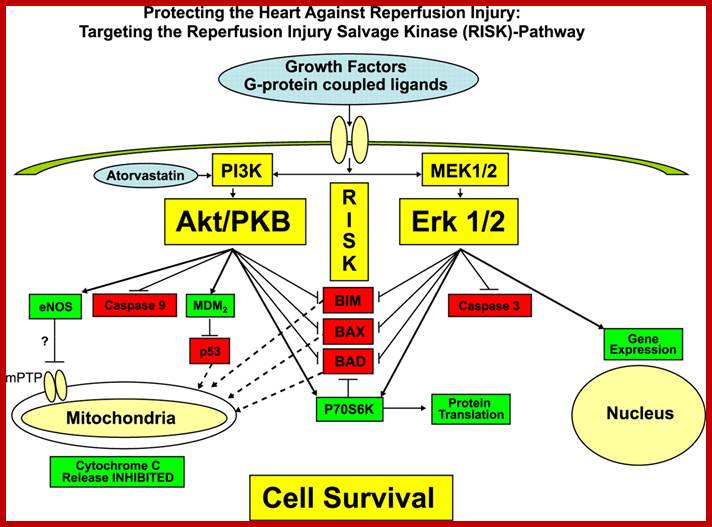
http://cardiovascres.oxfordjournals.org/
Irrespective of the morphological features of end-stage cell death (that may be apoptotic, necrotic, autophagic, or mitotic), mitochondrial membrane permeabilization (MMP) is frequently the decisive event that delimits the frontier between survival and death. Thus mitochondrial membranes constitute the battleground on which opposing signals combat to seal the cell's fate. Local players that determine the propensity to MMP include the pro- and antiapoptotic members of the Bcl-2 family, proteins from the mitochondrial permeability transition pore complex, as well as a plethora of interacting partners including mitochondrial lipids. Intermediate metabolites, redox processes, sphingolipids, ion gradients, transcription factors, as well as kinases and phosphatases link lethal and vital signals emanating from distinct subcellular compartments to mitochondria. Thus mitochondria integrate a variety of proapoptotic signals. Once MMP has been induced, it causes the release of catabolic hydrolases and activators of such enzymes (including those of caspases) from mitochondria. These catabolic enzymes as well as the cessation of the bioenergetic and redox functions of mitochondria finally lead to cell death, meaning that mitochondria coordinate the late stage of cellular demise. Pathological cell death induced by ischemia/reperfusion, intoxication with xenobiotic, neurodegenerative diseases, or viral infection also relies on MMP as a critical event. The inhibition of MMP constitutes an important strategy for the pharmaceutical prevention of unwarranted cell death. Conversely, induction of MMP in tumor cells constitutes the goal of anticancer chemotherapy.
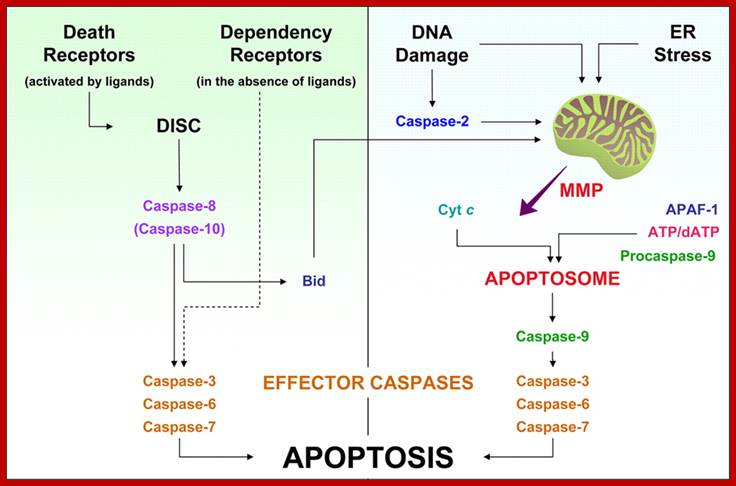
Extrinsic versus intrinsic caspase activation cascades. Left: extrinsic pathway. The ligand-induced activation of death receptors induces the assembly of the death-inducing signaling complex (DISC) on the cytoplasmic side of the plasma membrane. This promotes the activation of caspase-8 (and possibly of caspase-10), which in turn is able to cleave effector caspase-3, -6, and -7. Caspase-8 can also proteolytically activate Bid, which promotes mitochondrial membrane permeabilization (MMP) and represents the main link between the extrinsic and intrinsic apoptotic pathways. The extrinsic pathway includes also the dependency receptors, which deliver a death signal in the absence of their ligands, through yet unidentified mediators.Right: intrinsic pathway. Several intracellular signals, including DNA damage and endoplasmic reticulum (ER) stress, converge on mitochondria to induce MMP, which causes the release of proapoptotic factors from the intermembrane space (IMS). Among these, cytochrome c (Cyt c) induces the apoptosis protease-activating factor 1 (APAF-1) and ATP/dATP to assemble the Apoptosome, a molecular platform which promotes the proteolytic maturation of caspase-9. Active caspase-9, in turn, cleaves and activates the effector caspases, which finally lead to the apoptotic phenotype. DNA damage may signal also through the activation of caspase-2, which acts upstream mitochondria to favor MMP. See section IIA for further details. physrev.physiology.org
Operation of Death program:
Signal from outside:
On binding of signal molecules to their respective receptors (signal transducers) such as Fas-R and TNFa1-R, receptor undergoes conformational changes and gets activated.

When is the intrinsic pathway of apoptosis
activated? What signals are involved? Intrinsic- embryogenesis, hormone
induction (menstruation/menopause epithelium), injuries from radiation,
toxins, hypoxia; Increased mitochondrial permeability- release of cytochrome C.
Fas-ADD and caspase 8; http://quizlet.com/
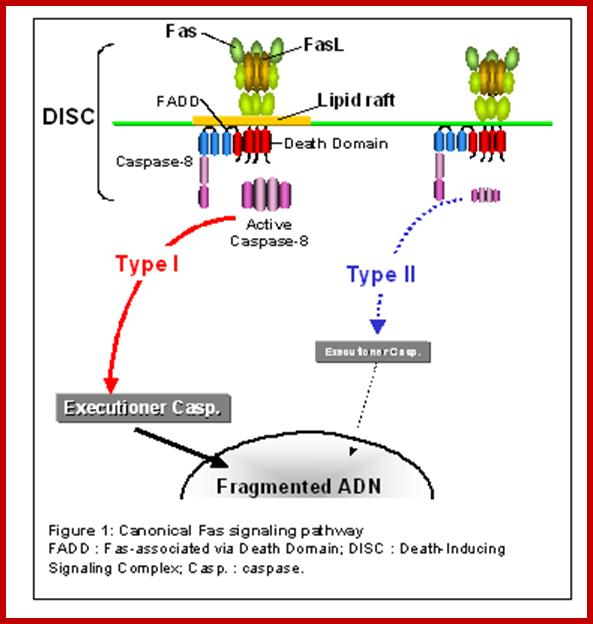
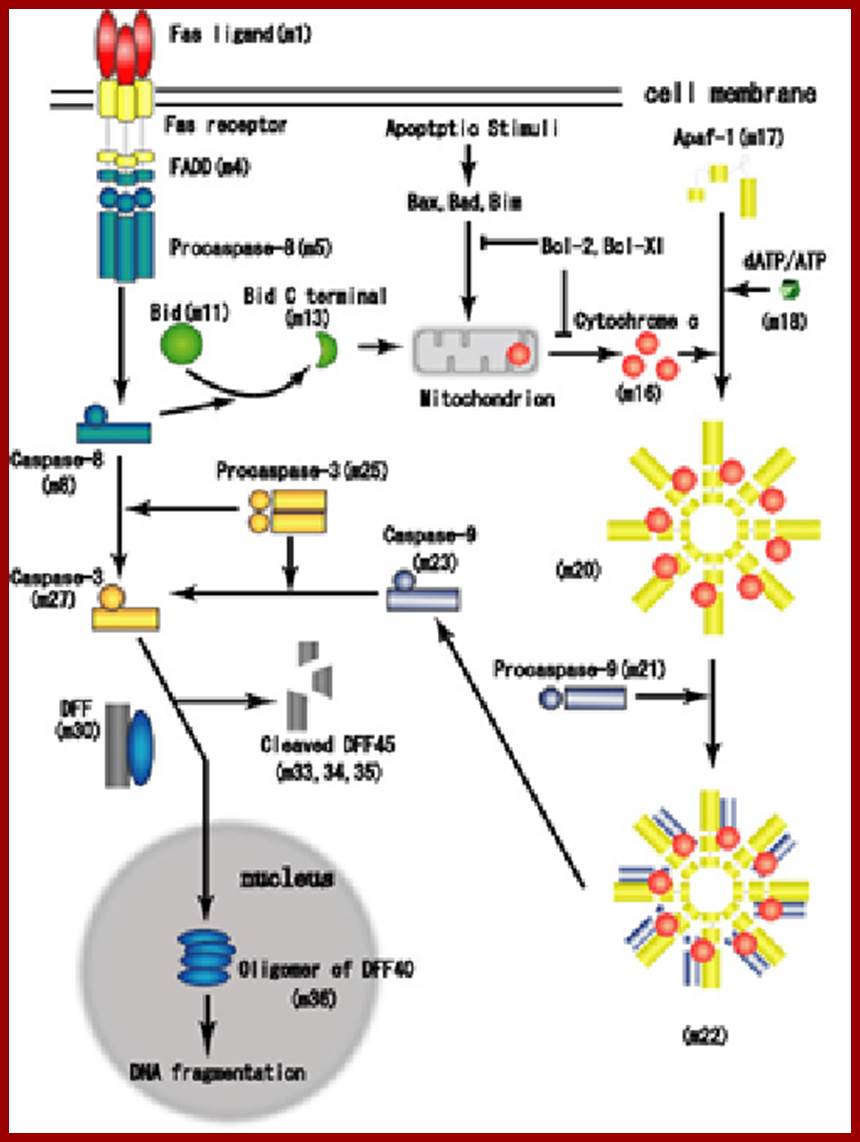
Proposed steps of apoptosis induced by Fas ligand: Fas ligands, which usually exist as trimmers, bind and activate their receptors by inducing receptor trimerization. Activated receptors recruit adaptor molecules such as Fas-associating protein with death domain (FADD), which recruit procaspase 8 to the receptor complex, where it undergoes autocatalytic activation. Activated caspase 8 activates caspase 3 through two pathways; The complex one is that caspase 8 cleaves Bcl-2 interacting protein (Bid) and its COOH-terminal part translocates to mitochondria where it triggers cytochrome c release. The released cytochrome c bind to apoplectic protease activating factor-1 (Apaf-1) together with dATP and procaspase 9 and activates caspase 9. The caspase 9 cleaves procaspase 3 and activates caspase 3. The other pathway is that caspase 8 cleaves procaspase3 directly and activates it. The caspase 3 cleaves DNA fragmentation factor (DFF) 45 in a heterodimeric factor of DFF40 and DFF45. Cleaved DFF45 dissociates from DFF40, inducing oligomerization of DFF40 that has DNase activity. The active DFF40 oligomer causes the internucleosomal DNA fragmentation, which is an apoptotic hallmark indicative of chromatin condensation.; Both the above diagrams show cascade of reactions leading the activation of Caspase and cell death; www. genomicobject.net

Cristae remodeling. Under physiological conditions, mitochondrial membranes define the boundaries of at least three sub mitochondrial compartments: the mitochondrial matrix is enclosed within the mitochondrial IM, the intermembrane space (IMS) is located between the IM and the mitochondrial OM, the intracristae space (ICS) is delimited by the convoluted folds of the IM, namely, cristae. Most cytochrome c (Cyt c) resides in the ICS, which communicates with IMS via tight bottleneck-like junctions, forming a diffusional barrier. It appears that ICS undergoes deep structural rearrangements during apoptosis to promote the complete release of Cyt c and other IMS proteins. In healthy cells, cristae structure is maintained with the help of Opa1 (optic atrophy 1) oligomers, which are constituted by both the IM integral form of Opa1 and by its IMS soluble counterpart. The serine protease PARL (presenilin-associated rhomboid-like) is responsible for the production of the soluble form of Opa1 that is required for the assembly of Opa1 oligomers. Upon apoptosis induction, Opa1 oligomers are disrupted (for instance following the translocation of truncated Bid, i.e., tBid). Then profound rearrangements of the sub mitochondrial structure take place, resulting also in the loss of the diffusion barrier between IMS and ICS. Taken together, these rearrangements have been called “cristae remodeling” and promote the mobilization of the pool of IMS proteins, including Cyt c, VI previously sequestered in ICS. Finally, mitochondrial membrane permeabilization (MMP) allows for the release of the mobilized IMS proteins. For further details see section; http://physrev.physiology.org/
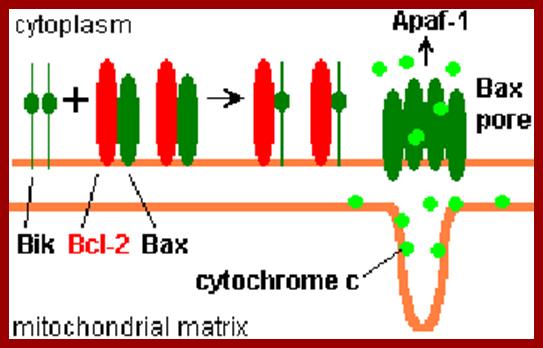
The above diagram depicts interaction of Bid with Bcl2-Bax transmembrane components located in the outer mitochondrial membrane leading to Bax pore formation and the release of Cyt.C. www.cell.com
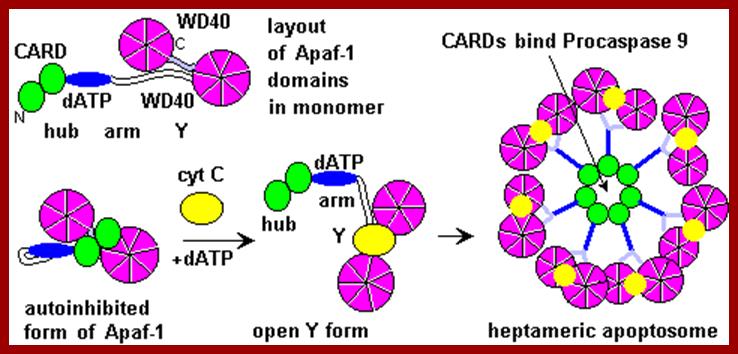
The figure above and the figure below shows the released Cyt.C binding to Apaf-1 at the domain called CARD, and further activated by the binding of ATP. This Y-complex binds to procaspase-9 and activates to cleave it and form a heptameric complex called Apoptosome, which in turn activates procaspase-3 that leads to devastating effect on cellular components and cell death. www.cell.com

Apaf-1-like molecules assemble into a ring-like platform known as the apoptosome. This cell death platform then activates procaspases in the intrinsic cell death pathway. In this review, crystal structures of Apaf-1 monomers and CED-4 dimers have been combined with apoptosome structures to provide insights into the assembly of cell death platforms in humans, nematodes, and flies. In humans, the caspase recognition domains (CARDs) of procaspase-9 and Apaf-1 interact with each other to form a CARD-CARD disk, which interacts with the platform to create an asymmetric proteolysis machine. The disk tethers multiple pc-9 catalytic domains to the platform to raise their local concentration, and this leads to zymogen activation. These findings have now set the stage for further studies of this critical activation process on the apoptosome. Apoptosome structure, Assembly and Procaspase Assembly, Shujun Yuan, Christopher W.Akey; www.cell.com
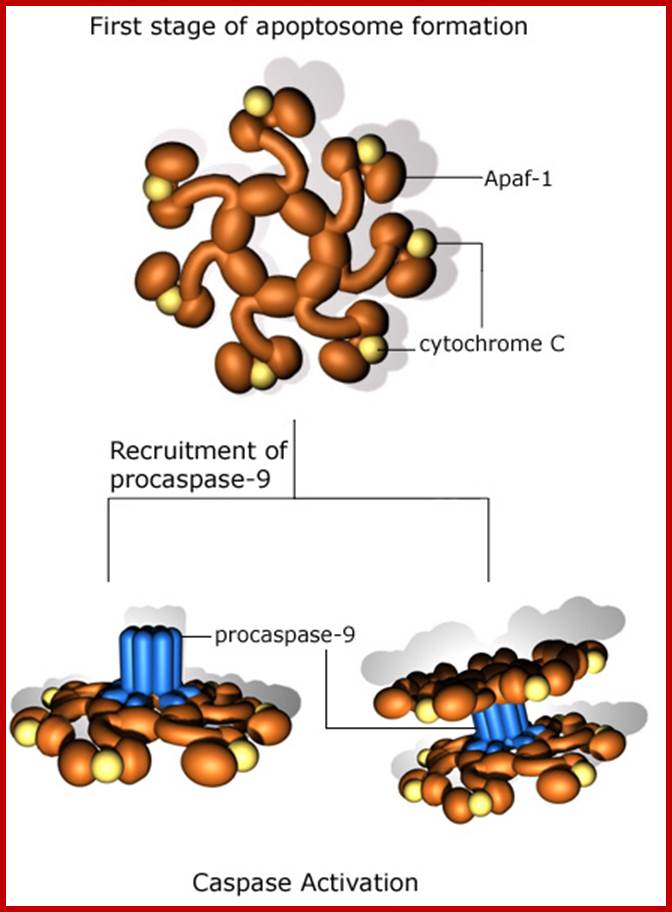
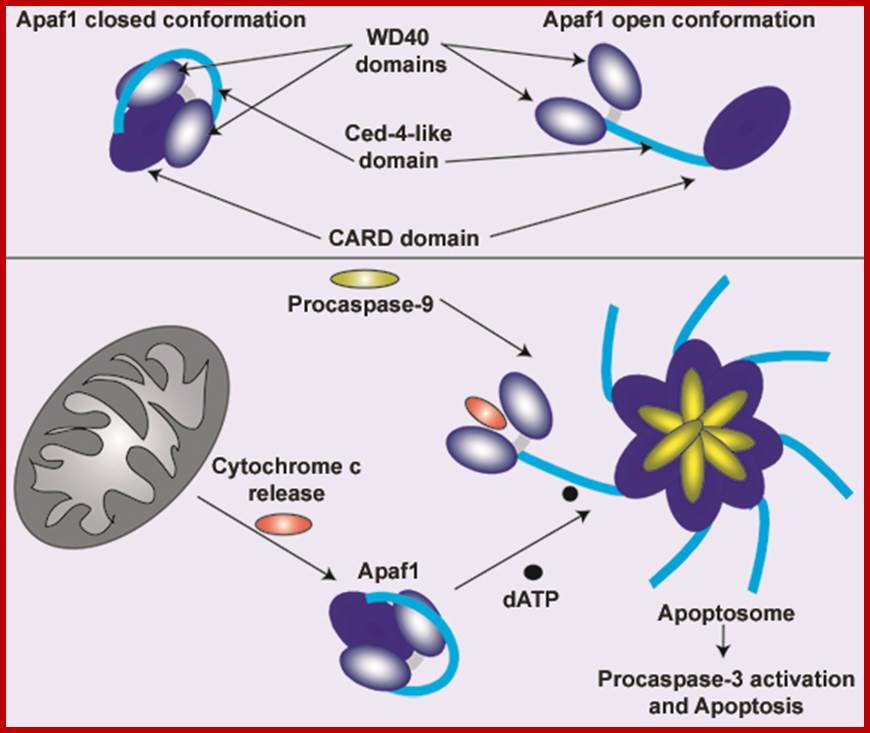
The conformational changes in the APAF1 molecules lead to apoptosome formation and to the activation of apoptosis. However, the assembly and the functioning of the apoptosome is regulated by mitochondrial and cytosolic factors (motif. from E. Ferraro et al. 2004) www.atlasgeneticsoncology.org
Apoptosis triggered by external signals: the extrinsic or death receptor pathway
- Fas and the TNF receptor are integral membrane proteins with their receptor domains exposed at the surface of the cell
- binding of the complementary death activator (FasL and TNF respectively) transmits a signal to the cytoplasm that leads to
- activation of caspase 8
- caspase 8 (like caspase 9) initiates a cascade of caspase activation leading to
- phagocytosis of the cell.
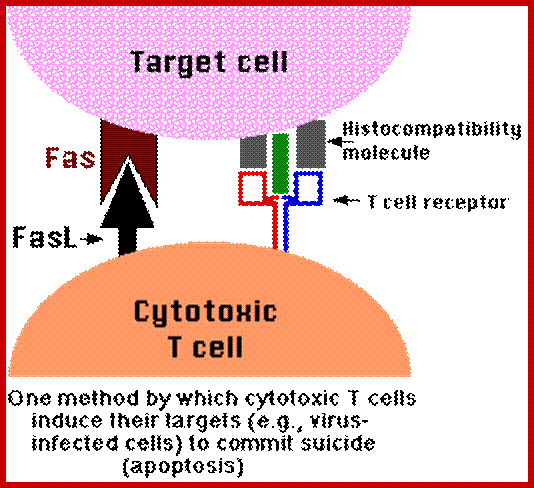
Example (right): When cytotoxic T cells recognize (bind to) their target,
- They produce more FasL at their surface.
- This binds with the Fas on the surface of the target cell leading to its death by apoptosis.
The early steps in apoptosis are reversible — at least in C. elegans. In some cases, final destruction
Apoptosis-Inducing Factor (AIF):
Neurons, and perhaps other cells, have another way to self-destruct that — unlike the two paths described above — does not use caspases.
Apoptosis-inducing factor (AIF) is a protein that is normally located in the intermembrane space of mitochondria. When the cell receives a signal telling it that it is time to die, AIF
- is released from the mitochondria (like the release of cytochrome c in the first pathway);
- migrates into the nucleus;
- binds to DNA, which
- triggers the destruction of the DNA and cell death (http://users.rcn.com/jkimball.ma.ultranet/
First they cluster as trimers and more clusters develop. The activated cytosolic domain now binds with adaptor proteins such as FADD in the case of Fas 1-R and TRADD and FADD in the case of TNF a1-R. All are in active state. At this juncture cellular precursor Casp-8 interacts with the adaptor proteins through its own death domains, this complex is called death inducing signal complex (DISC), which in turn this activates, casp-8. The activated casp-8 performs auto cleavage of its own structure into a large and small subunit and they bind to each other as dimers proteins.
Activated can directly act on casp-3 and activates the death process. The second route it takes is to act on Bid to activate mitochondrial mediated process.
This activated casp-8 (as dimeric protein) now acts on Bid protein and cleaves into N-end and C-end subunits. The c-end subunit translocates into outer mitochondrial membrane and interacts with Bax proteins. Found in the mitochondrial membranes. In the membrane the Bax proteins are associated with another class of proteins called Bcl2. If the Bcl2 exist as homodimers, interaction of Bid fails to release Cyt-C from the periplasmic space of Mitochondria. But on the contrary if the Bax is associated with Bcl2 as heterodimers, they facilitate the release of Cyt-C into cytosol. The concentration of Bcl2 and Bax is very important. If the Bax proteins are in homodimeric state, Cyt-c will be released. If the Bcl2 are in homodimeric state, Cyt-c is not released, but if the Bax and Bcl2 are in heterodimer state, the Bid interaction induces them to release Cyt-C. The Bcl2 protein when expressed in higher amounts, it induces prevents apoptosis and induces cell proliferation.
The released Cyt-C interacts with pro-casp-9 and activates it through another protein called ApaF1. First Cyt-c binds to ApaF1 and this dimer binds to ATP. The activated dimers interact with pro-Casp-9 and cleave it to active casp-9. This complex of Cyt-c, ApaF1 and casp-9 is called Apoptosome. Such trimers can interact with one another and form a ring of huge Apoptosome complex.
The Apoptosome acts on pro-casp-3 (ced3). The casp-9 also activates casp-6 and 7. The activated casp-3 activates some downstream proteins such as Caspases activated DNAase (CAD). This DNAase is found in cytoplasm, but an inhibitor of CAD called ICAD binds it. Caspase-3 degrades the ICAD and releases the DNAase in active form, which enters into the nucleus and cleaves the DNA. Though there are Inhibitors of apoptosis (IAPs) within the cell, they are activated casps. Diablo inactivates IAPs. Mitochondrial also releases endonuclease G that enters the nucleus and cleaves DNA into 180 bp long fragments. Ced-3 virtually dismembers all cellular protein especially of microtubules and other cytoskeleton proteins. This act of enzyme causes blebbing and destruction of cells.
Signal from inside:
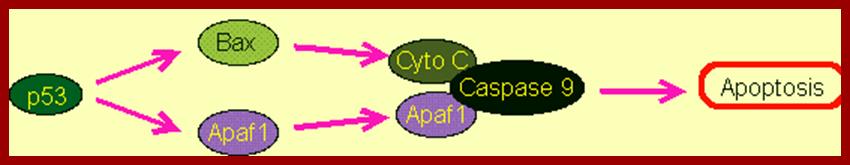
When the cellular DNA is damaged because of errors in replication or damaged by irradiations, or due to the loss of protection of Telomeric DNA, leads to fragmentation of DNA with 3’hanging ends. Such fragments are recognized by p53 proteins through Rad9 and the p53 gets activated via phosphorylation by Akt members. P53 proteins are considered as tumor suppressors. Binding of p53 to DNA ends activates the protein, the activated tetramers activate the transcription of certain genes, and one such is the gene for p21 and p27 proteins. The p21 and p27 proteins inhibit the activity of Cdk-cyclin complexes and hold the cell at G1-S transitional stage. The activated p53, if finds the DNA damage is severe the p53 by its N-terminal domain interacts with mitochondrial membranes and promotes the release of Cyt-C, which activates the whole process of cell destruction. It is for this reason the N-terminal region of p53 is called apoptotic domain. The actual apoptotic domain lies within the first 63 amino acid residues from the N-end; especially residues 53 and 54 are critical for both apoptotic and transcriptional activation.

The p53 pathway. Awakening guardian angels: drugging the p53 pathway;
Christopher J. Brown, Sonia Lain, Chandra S. Verma, Alan R. Fersht & David P. Lane
p53 is at the centre of a complex web of biological interactions that translates stress signals into cell cycle arrest or apoptosis26. Upstream signalling to p53 increases its level and activates its function as a transcription factor in response to a wide variety of stresses, whereas downstream components execute the appropriate cellular response. The principal sensors seem to be MDM2 and MDM4 and their interaction with p53. In non-stressed conditions these proteins bind p53, ubiquitylate it and target it for degradation by the proteasome. In stressed conditions the function of the MDM2–MDM4 complex is blocked by phosphorylation, protein-binding events and/or enhanced degradation141. Hence, phosphorylation of MDM4 is essential for the p53 response to ionizing radiation, and the response to oncogene activation depends on the binding of ARF to MDM2. Many p53-activating small molecules function by causing the release of ribosomal proteins from the nucleolus to the nucleoplasm, where they bind to MDM2 and MDM4 and inhibit their function. Molecules that activate wild-type p53 in tumours by disrupting MDM2 activity can compensate for any missing upstream components of the p53 pathway, for example the loss of ARF expression that is frequent in cancer cells142. However, defective downstream p53 signalling might substantially decrease their effectiveness. Therefore, the ability to identify tumours in which downstream p53 signalling is unaffected is important. The development of strategies to ensure that the desired p53 response is initiated when it is reactivated might be necessary and could require the judicious use of drug combinations. 53BP1, p53 binding protein 1; ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3 related; BAI1, brain-specific angiogenesis inhibitor 1; BAX, BCL2-associated X protein; BBC3, BCL2 binding component 3 (also known as PUMA); DR, death receptor; GADD45, growth arrest and DNA-damage-inducible 45; KILLER, p53-regulated DNA damage-inducible cell death receptor (also known as TNFRSF10B); LRDD, leucine-rich repeats and death domain containing; miRNA, microRNA; PMAIP1, phorbol-12-myristate-13-acetate-induced protein 1 (also known as NOXA); RPRM, reprimo; RRM2B, ribonucleotide reductase M2 B; ST13, suppression of tumorigenicity 13 (also known as p48); TP53I3, tumour protein p53 inducible protein 3; THBS1, thrombospondin 1; UV, ultraviolet.
Other pathways:
1. Fas can also be activated to execute apoptotic pathway by using JNK kinase, whose substrate is C-Jun a transcription factor. This pathway activates certain set of proteases. When the signals are impinged upon the Fas-R, the activated receptor proteins interact at cytosolic side with DaXX. The TNF- receptor can also be activating JNK pathway by another distinct adaptor proteins, however the stress related activation of JNK is independent of Fas pathway, which is not inhibited by Bcl2. The intermediate reaction in JNK pathway starts from Fas-R/TNF-R. The activated Fas-R/TNF-R binds to DaXX at cytosolic surface. They in turn interact with JNK system, which then phosphorylates C-Jun. The phosphorylated c-Jun. is involved in production of Caspases that leads to apoptosis.
2. In the absence of death inducing signals, where cells are destined to die, cells do die by their own self-suicidal mechanism. Do human beings have that mechanism? When the cell is aged and the cell death receptor not activated or in the absence of Trophic factors, a cytosolic protein called Bad (Bcl-xl/Bcl2 associated X-protein), associates Bcl2-BclX1 proteins found at the outer membrane of mitochondria and activate the transport system to release Cyt.C., that activates a cascade of reaction leading to the death of the cells.
The fascinating aspect of this phenomenon is, in presence of a mitogenic factor, when the ligand binds, the cytosolic domain activates Phosphotidyl inositol-3 kinase) PI-3. The activated PI-3 activates AKT kinase, which is another one of the protein kinases, phosphorylates Bad protein. The phosphorylated Bad proteins are sequestered by a an un-usual protein called 14.3.3. this blocks the release of Cyt.C. thus blocks cell death by suicide.
Molecular mechanisms of caspase regulation during apoptosis;
Caspases, which are the executioners of apoptosis, comprise two distinct classes, the initiators and the effectors. Although general structural features are shared between the initiator and the effector caspases, their activation, inhibition and release of inhibition are differentially regulated. Biochemical and structural studies have led to important advances in understanding the underlying molecular mechanisms of caspase regulation. This article reviews these latest advances and describes our present understanding of caspase regulation during apoptosis.
T-Cell mediated cytotoxic pathway:
In the case of cytotoxic mediated cell death, specific cellular proteins at their surface mark the cells. The activated cytotoxic T-lymphocytes bind to such surface proteins and kill their target cells by releasing Granzymes in the form of granules. Perforin creates holes in the receiver cells, thus allow all the enzymes to enter into the target cell. Granzyme-B induces many features similar to apoptosis including DNA fragmentation. It activates Caspase-3, which is very essential for apoptosis.
Perforin/Granzyme Killing
CTLs have cytoplasmic granules that contain the proteins perforin and granzymes. When the CTL binds to its target, the contents of the granules are discharged by exocytosis.
A dozen or more perforin molecules insert themselves into the plasma membrane of target cells forming a pore that enables Granzymes to enter the cell. Granzymes are serine proteases. The two most abundant ones are
- Granzyme A. Once inside the cell, it enters the mitochondria and cleaves a subunit of complex I (the NADH dehydrogenase) of the electron transport chain producing reactive oxygen species (ROS) that kill the cell.
- Granzyme B. Once inside the cell, it proceeds to cleave the precursors of Caspases thus activating them to cause the cell to self-destruct by apoptosis.
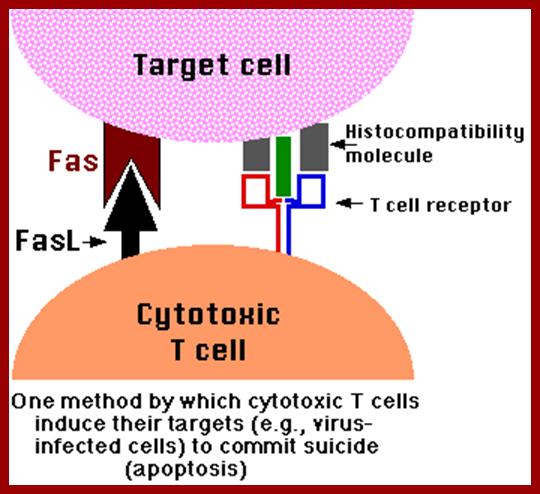
FasL/Fas Killing; http://users.rcn.com/
CTLs express on their surface molecules of a transmembrane protein, the death activator designatedFas ligand (FasL). Most potential CTL targets express a receptor for FasL designated Fas. When cytotoxic T cells recognize (bind to) their target, they produce more FasL at their surface. This binds with the Fas on the surface of the target cell leading to its death by apoptosis.;users.rcn.comusers.rcn.com
By using this technology, it is possible to destroy tumor cells by a process called cell mediated therapy. Tumors contain their own lymphocytes called Tumor infiltrated lymphocytes called TILs. Such TILs can be recovered and activated by specific interleukins. The activated TILs become killer cells. When such activated TILs are injected into the patient TILs reach their specific tumors and by binding they release Granzymes and kill the targets cells, thus it is possible to remove tumors.
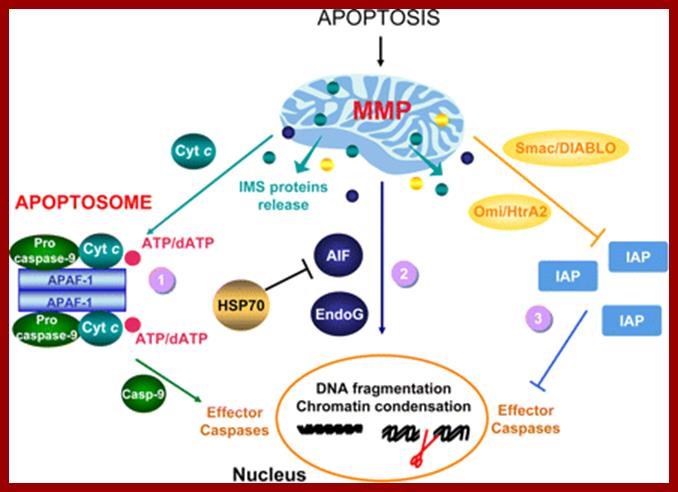
Release of IMS proteins. Proapoptotic signals resulting in mitochondrial membrane permeabilization (MMP) provide intermembrane space (IMS) proteins with a route for release. Once in the cytosol, IMS proteins follow different fates. 1) Cytochrome c (Cyt c) promotes the formation of the so-called “apoptosome,” a molecular platform for the activation of caspase-9 (Casp-9) including also the apoptosis protease activating factor 1 (APAF-1) and ATP/dATP. In turn, active Casp-9 catalyzes the proteolytic activation of the effector caspases, which ultimately contribute to the appearance of the morphological hallmarks of apoptosis (e.g., DNA fragmentation and chromatin condensation). See sections IIA and VIIIA as well as Figure 1for further information. 2) The caspase-independent death effectors apoptosis-inducing factor (AIF) and endonuclease G (EndoG) translocate from the cytosol to the nuclear compartment where they favor DNA fragmentation and chromatin condensation. Members of the heat shock protein (HSP) family, like HSP70 (i.e., heat shock protein of 70 kDa), antagonize AIF proapoptotic activity by preventing its nuclear import. For additional details, see sections VIII, C and D, as well as Figure 11. 3) Second mitochondria-derived activator of caspase/direct IAP binding protein with a low pI (Smac/DIABLO) and the Omi stress-regulated endoprotease/high temperature requirement protein A2 (Omi/HtrA2), promote apoptosis indirectly, by binding to and antagonizing members of the IAP (inhibitor of apoptosis protein) family. Under normal circumstances, IAPs would exert antiapoptotic effects by preventing the caspase activation. See section VIIIB for more detailed information.www. http://physrev.physiology.org/
Apoptosis vs Carcinogenesis:
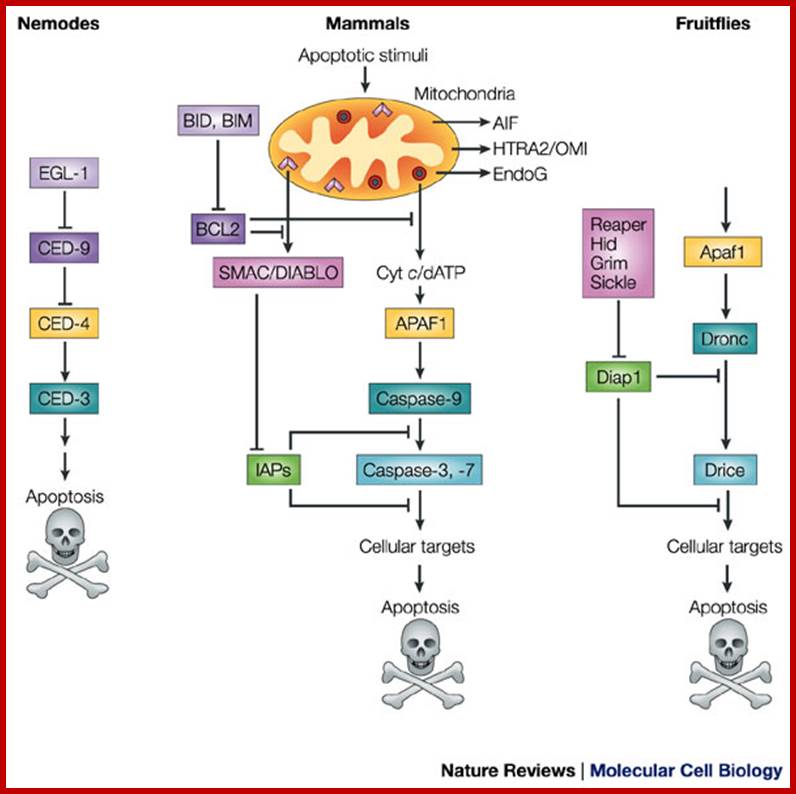
Genetic studies in C.elegans have revealed programmed death of those 130 cells among 1090 cells is due to certain genes. Some of them are pro-apoptotic and some of them are anti-apoptotic. Similar genes and more have been identified in human systems. Gene knockout experiments using cultured transgenic cells, the role of each of the genes has been elucidated to some extent. In C.elegans the genes CED3 and CED4 leads to cellular death and CED-9 prevents cell death. The counterparts in human system are Bcl-2, Apaf-1, Casp9 and Casp3; the first one Bcl2 is a suppressor of apoptosis and the remaining three are pro-apoptosis factors.
C.elegans: Ced-4-àCed-3-àCell death, this path way is blocked by Ced-9.
Human: Apaf-1-à Casp-9 -à Casp3 -àcell death; this pathway is blocked by Bcl-2.
Expression of CED4 in kidney cells leads to rapid apoptosis. This can be blocked by the co-expression of Bcl2 or CED9. CED4 binds to CED3 and activates protease activity that leads to the degradation of cellular proteins, hence cell death. Bcl2 and CED9 are homologous proteins found on the outer membrane of mitochondria, endothelial membranes and outer nuclear membranes. These proteins have a single transmembrane domain
The key cellular component responsible for destruction of cellular proteins is Caspase. It is a cysteine-protease when activated, targets proteins importantly cellular microtubules and nuclear lamins are attacked and degraded by cutting the peptide bond towards the carboxyl side of the Aspartate amino acid; that is the reason why nucleus and cells collapse and crumble. In C.elegans the most effective molecule is CED3. In mammals they have multiple Caspase. CED9 binds to CED4 and localizes from the cytosol to cellular membranes. So CED’s apoptotic function is suppressed but CED4 binds to CED3 and activates protease activity.
Another pro-apoptotic factor is Bax, which is found associated or complexed with Bcl2, but it’s over expression causes apoptosis. Sequence of Bax protein is similar to that of CED9 and Bcl2, but over expressing Bax induces death of cells. All these proteins have single transmembrane domains and enable oligomerization.
Bcl2 belongs to a family of proteins involved in distribution of Cytochrome-C, which in normal conditions found in the space between outer and inner mitochondrial membranes; actually this is a marker protein. Bcl2 blocks the release of this protein, but Bax counteracts the Bcl2 and promotes the release of Cyt.C into cytoplasm. The released Cyt.C binds to apoptosis activation factor-ApaF-1 (CED-4) and activates Caspase cascade.
The release of Cyt.C from the mitochondrial membranes into the cytosol is due to the influx of ions from the cytosol, this due to the binding of homo-dimers of Bax but not Bcl2/Bax heterodimers to mitochondrial membranes. Bcl-xl is another anti-apoptotic protein. When this gene is knocked out of mice there is a massive cell death in spinal cord and brain regions, but knock out of Bax leads to marked increased neuronal tissues.
Trophic factors work independent of protein synthesis. Binding of Trophic factors to their respective receptors induce signal transduction and down stream signaling process where certain proteins are post translationally modified. The Bad protein in it non-phosphorylated form associates with Bcl2/Bcl-xl at mitochondrial membranes. Binding of Bad (-P) to Bcl2/Bcl-xl inhibits anti-apoptotic function of Bcl2/Bcl-xl. But phosphorylated Bad (+P) fail to bind to Bcl2/Bcl-xl, for phosphorylation leads to sequestering of bcl2/Bcl-xl by phosphoserine binding proteins in cytosol.
Certain growth factors activate PI-3 kinase, which in turn can activate downstream kinase such as Akt. This kinase phosphorylates Bad at specific sites; this is known to inhibit pro-apoptotic activities. Constitutively active Akts can rescue cultured cells that are deprived of neutrophin from death.
As in the case of cell proliferation, mitogen factors stimulate cell proliferation; if it is not controlled it leads to cancer. Similarly certain damage to genetic material in the cells, which is beyond repair system to normality, certain molecular sensors with in the cells, sensing the severity of damage; trigger the process of cell death, called Apoptosis.
There is a relationship between tumor formation and Apoptosis. P53 activates several genes involved in cell cycle regulation. ADV’s E1B, a 19KD protein blocks P53 ‘s transcriptional activation. Bcl2 is an oncogene identified for its activity in tumors. It actually blocks the pathway of Apoptosis. So Apoptosis inhibits tumorigenesis and Bcl2 prevents Apoptosis. P53 has pleotropic phenotypic effects; it triggers growth arrest if the DNA damaged and induces Apoptosis if the damage is beyond repair. RB and P53 are activated in multiple routes. One locus that influences both RB and P53 is INK4A-ARF. Its transcript is alternately spiced as P16NK4A in RB and p19ARF in P53.
In human cancers, deletion of INK4A-ARF is common which eliminates both p16INK4A and P19ARF, thus cell loses the ability to block tumor formation. P16INK4A inhibits cdk4/6 kinase, so it prevents the kinase from phosphorylation RB. In the absence of it cell cycle progression is halted in its track. The activity of INK4A is often disabled because of several point mutations in human tumors. P19ARF antagonizes Mdm2, which leads to stabilization of p53. Thus p19ARF acts as a tumor suppressor by inhibiting the inhibitor of P53. P19 may promote the degradation of Mdm2 or directly blocks interaction with p53. p19ARF arrests the cell cycle in p53 dependent manner. Loss of p19ARF and p53 has the same effect.
C-myc and E1 (ADV) act via p19ARF to activate p53 dependent activation. P53 is often phosphorylated at serine residues (S6, S9, and S37etc). P53 is also acetylated at different positions. Ionizing radiations activate the kinase ATM, which phosphorylates S13 and S33 and S376 and L382. Such modifications may affect P53 ‘s stability, oligomerization, DNA binding and the ability to bind to other proteins. So p53 acts as a molecular sensor.
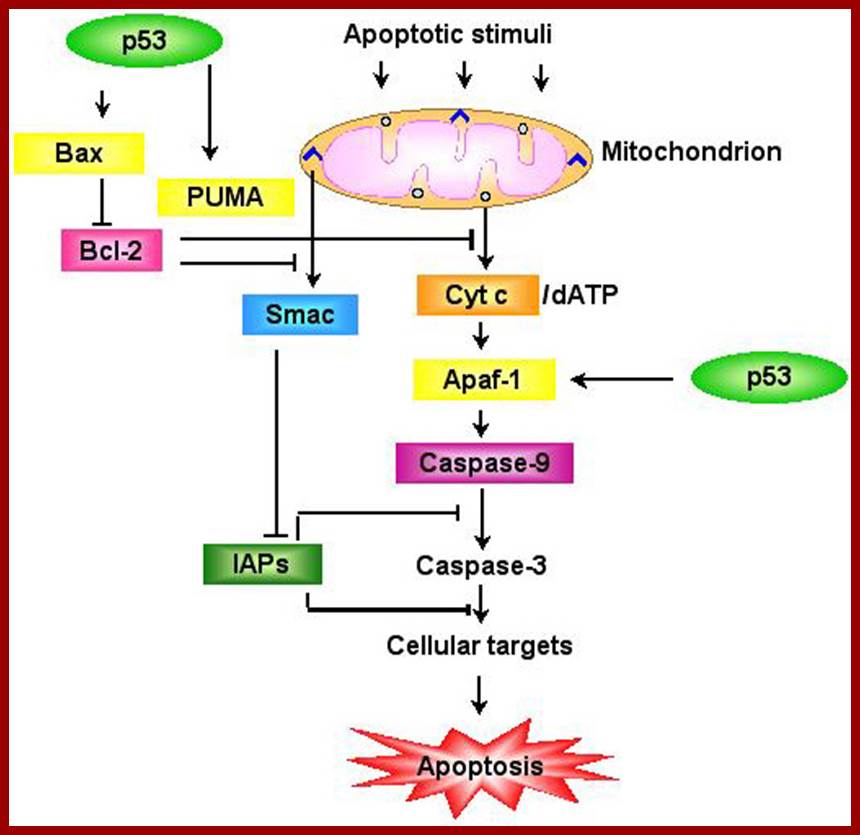
Schematic representation of the p53-dependent apoptotic pathways by transcriptional activation of BAX, PUMA and APAF-1.;www.weizmann.ac.
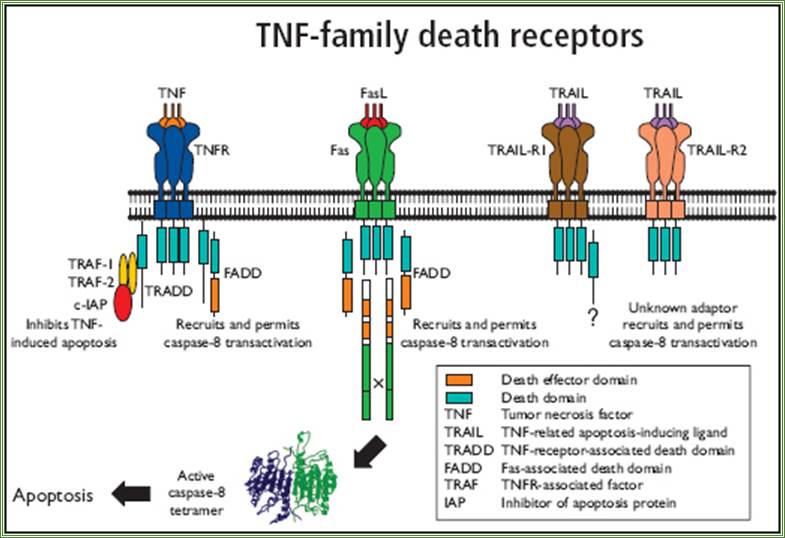
Extrinsic or Death Receptor Apoptosis Pathway
The extrinsic or death receptor apoptosis cell signalling pathway is initiated through ligand binding to death receptors on the cell membrane. The death receptors TNF-R1 (CD120a), TNF-R2 CD120b), Fas/APO-1 (CD95) and the TNF-related apoptosis-inducing ligand (TRAIL) receptors called DR3, DR4, DR5 and DR6 are subgrouped under the tumor necrosis factor (TNF) receptor superfamily.
This family also includes TWEAK Receptor (CD266 / Fn14) which interacts with its ligand TWEAK (TNF-like weak inducer of apoptosis) (CD255) inducing apoptosis in some tumour cell lines through pathways yet to be fully elucidated. http://www.abdserotec.com/
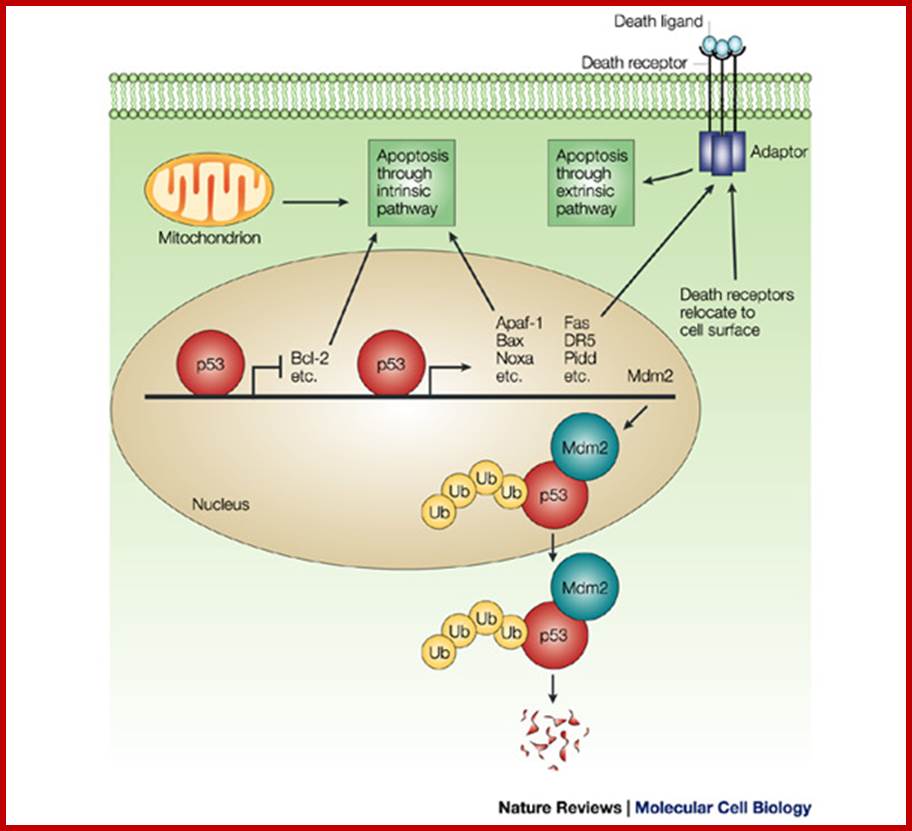
p53 induces the expression of proteins that target both the mitochondrial- and the death-receptor-induced apoptotic pathways, and specifically represses transcription from several death-inhibiting genes. Further activities of p53 that are entirely independent of transcriptional regulation have been proposed. They include the ability of p53 to drive relocalization of death receptors such as Fas/CD95 from the Golgi to the cell surface and to directly associate with mitochondria. Central to the regulation of p53 is Murine double minute 2 (Mdm2), which itself is a transcriptional target of p53. Mdm2 binds to p53 and targets p53 for ubiquitin/proteasome-dependent degradation. Ubiquitylation (Ub) of p53 by Mdm2 probably also enhances the export of p53 from the nucleus to the cytoplasm, where degradation takes place. Bcl-2, B-cell lymphoma 2; Apaf, Apoptotic protease-activating-factor; Bax, Bcl-2 associated X protein; DR5, death receptor 5; Pidd, p53 protein induced, with death domain. Veronika Jesenberger and Stefan Jentsch; http://www.nature.com/
p53 tumour suppression; Alex N. Bullock & Alan R. Fersht
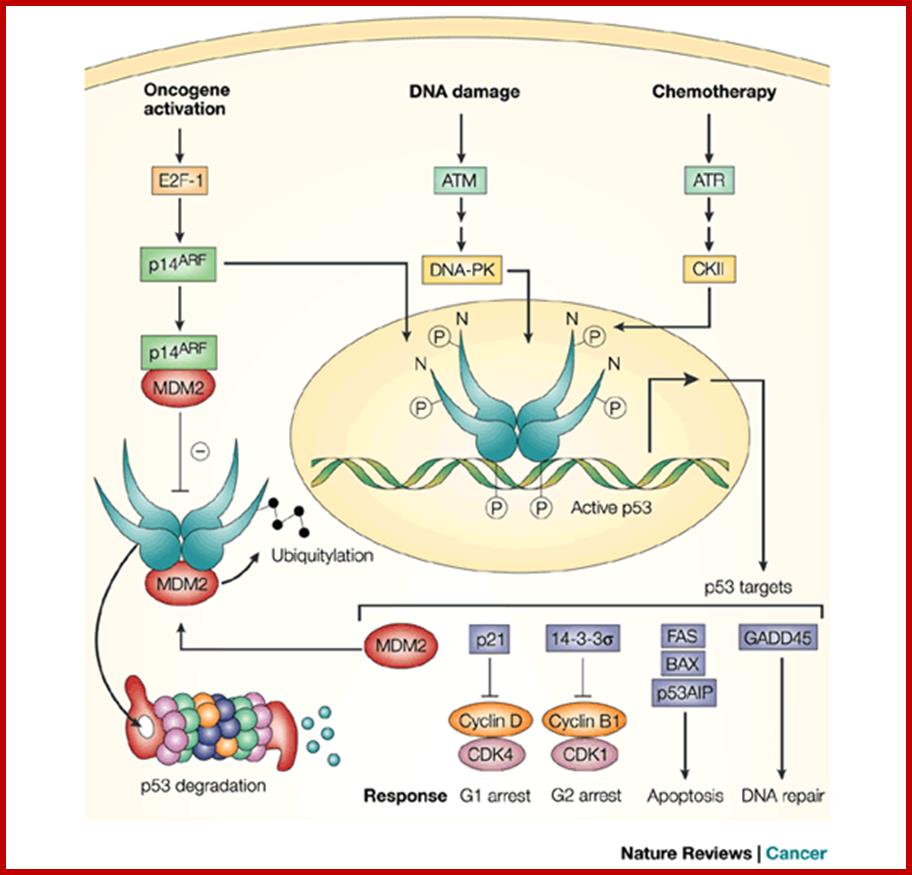
A
negative regulatory feedback loop controls cellular level of p53. In normal
cells, p53-dependent transcription of MDM2 promotes
p53 degradation. Cellular stress, such as oncogene activation, induces p14ARF,
which sequesters MDM2. In addition, DNA damage and chemotherapeutic agents
activate protein kinases, such as ATM and ATR, which, through DNA-dependent
protein kinase (DNA-PK) and casein kinase II (CKII), respectively,
phosphorylate the amino terminus of p53 to prevent MDM2 binding, and the
carboxyl terminus of p53 to increase sequence-specific DNA binding. These
events increase p53 levels and activate the transcription of p53 target genes.
p21 and 14-3-3![]() promote growth arrest at the G1 and G2 DNA-damage
checkpoints by inhibiting cyclin-dependent protein kinase (CDK) activity; FAS,
BAX and p53AIP promote apoptosis if repair is not possible; and GADD45 promotes
DNA repair. p53 also has poorly characterized roles in anti-angiogenic pathways
(not shown).
promote growth arrest at the G1 and G2 DNA-damage
checkpoints by inhibiting cyclin-dependent protein kinase (CDK) activity; FAS,
BAX and p53AIP promote apoptosis if repair is not possible; and GADD45 promotes
DNA repair. p53 also has poorly characterized roles in anti-angiogenic pathways
(not shown).
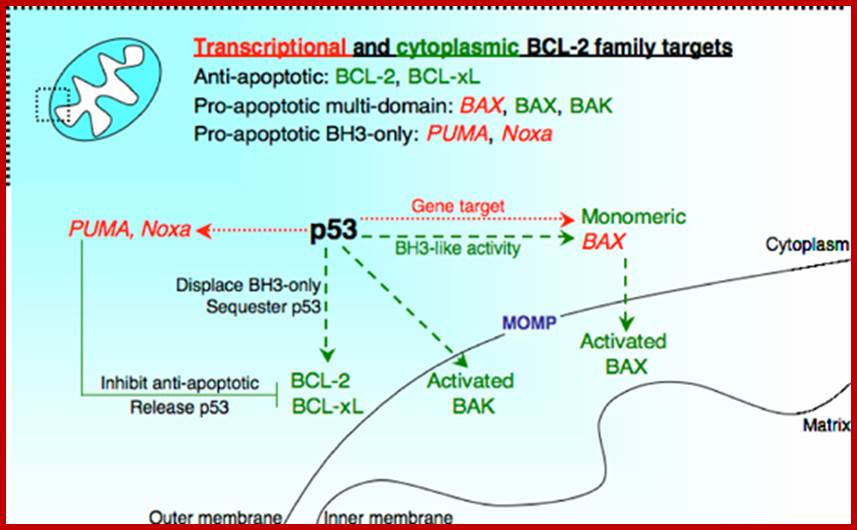
The relationship between p53 and Bcl-2 proteins at the mitochondrial membrane. p53 can either induce the expression of pro-apoptotic Bcl-2 proteins (e.g., Bax, Puma, Noxa), or it can directly regulate numerous Bcl-2 proteins in the cytoplasm. Bax that is expressed by p53 can then be activated by numerous signals, such as p53-induced BH3-only proteins (such as tBid), or by p53 itself. Bax or Bak that is activated by p53 or a BH3-only protein oligomerizes in the mitochondrial outer membrane to allow for MOMP. When Bcl-xL or Bcl-2 is bound by p53, BH3-only proteins once associated with either anti-apoptotic molecule may then be released to either directly activate a pro-apoptotic multi-domain or 'inhibit' an additional anti-apoptotic Bcl-2 protein. If nuclear p53 induces the expression of additional BH3-only proteins when cytoplasmic p53 is associated with Bcl-xL, the cell may be sensitized to numerous inducers of death. Alternatively, p53 that is sequestered by Bcl-xL may be liberated by a collateral signal, such as Puma, which frees p53 to activate Bax. Red and green arrows indicate transcriptional and cytoplasmic regulation, respectively; http://www.nature.com/
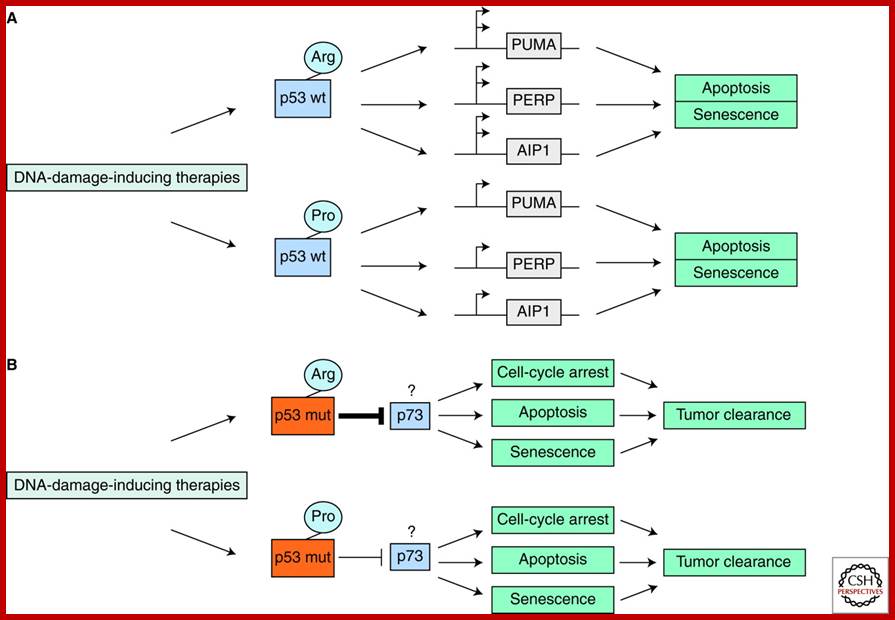
Single-nucleotide Polymorphisms in the p53 Signaling Pathway; Incorporating information of both the inherited and somatic genetics of the p53 gene could further define patient populations in their abilities to respond to certain therapies. (A) Some studies suggest that cells from individuals with the proline (Pro) allele of p53 codon72 will undergo less apoptosis in response to DNA-damage-inducing therapies compared with individuals with the arginine (Arg) allele of p53 codon72. This has been suggested to be caused by less transcriptional activation of apoptotic effectors. (B) Other studies suggest that cancer cells with somatic p53 mutations from individuals with the proline (Pro) allele of p53 codon72 will undergo more apoptosis in response to DNA-damage-inducing therapies compared with individuals with the arginine (Arg) allele of p53 codon72. This has been suggested to be potentially because of an enhanced inhibition of the p73 tumor suppressor by mtp53-codon72-Arg. http://www.cshperspectives.com/
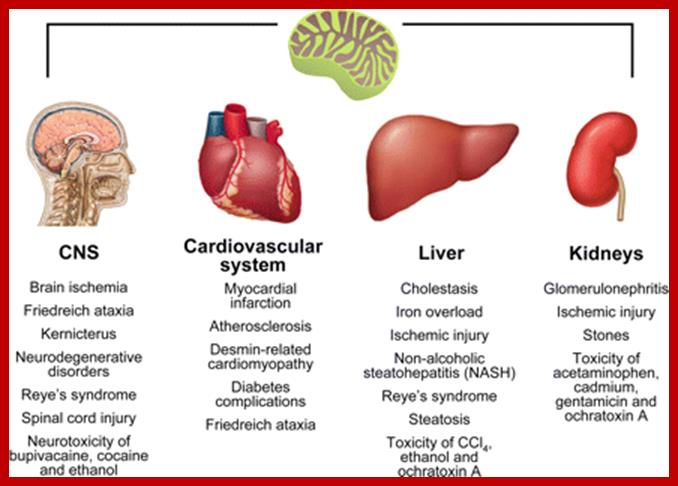
Examples of the involvement of mitochondrial apoptosis in pathological cell loss. Mitochondrial apoptosis has been implicated in a plethora of acute and chronic human diseases that affect several tissues and organs. The figure reports only a few examples of pathological conditions in which mitochondrial apoptosis plays a prominent role, grouped according to the most affected tissue. Please refer to section IX for further details. CNS, central nervous system; MMP, mitochondrial membrane permeabilization. www. http://physrev.physiology.org/
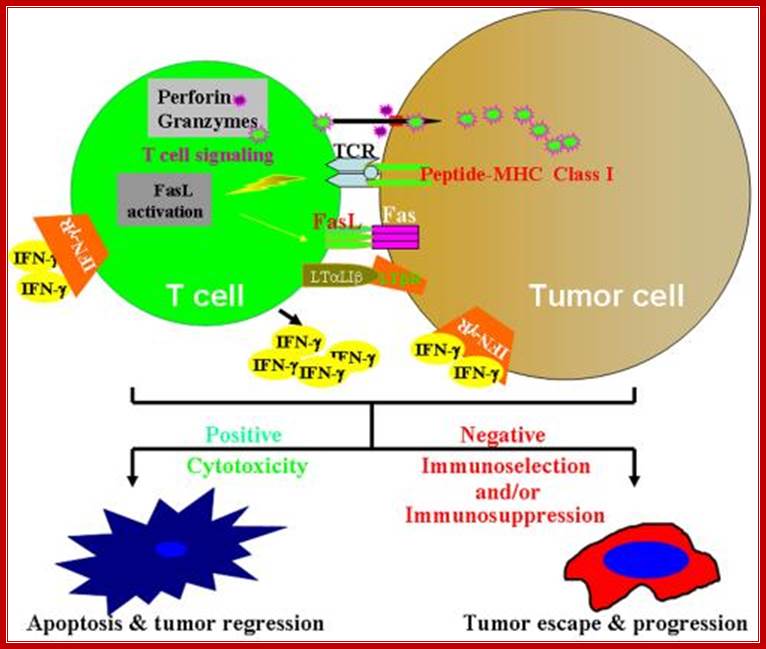
Model of immune cell and tumor cell interactions: when tumor cells arise in normal tissue, an inflammatory response is induced which attracts immune cells migrating to the tumor site. Interactions between immune cells and tumor cells involve direct cell-cell physical contact and release of modulator molecules (i.e. IFN-ý, TNFα and many other molecules). In this life and death battle, on the one hand, the immune cells may prevail and eradicate the tumor. On the other hand, tumor cells can counterattack the immune cells by producing inhibitory molecules, manipulating the immune suppressive cells, or acquiring resistant mechanisms to avoid induction of apoptosis by the immune cells. Thus, targeting cancer cell escape mechanisms is essential for effective cancer immunotherapy. http://www.gru.edu/
Some Acronyms n Expansions:
APAF1 = Apoptotic protease activating factor.
Apoptosome: Apf1-Cyt-C, dATP-Pro Caspase9-ced 4 complex.
BAD=BCL2 antagonist of cell death.
BAX=BCL2 associated X protein.
BCL2= B-cell Cell lymphoma 2.
Bcl X = B cell cell lymphoma factor that inhibits cell death.
BID= BH3 interacting domain death agonist.
Ced = Cell death genes, 1--XXX
DAXX= death associated protein 6.
DR = Death receptors.
FADD= Fas (TNFRSF^) associated death domain.
FAS = CD95, and Apo 1.
FASL = Fas ligand
TRADD= TNFRSF1A-associated death domain.
Ces= cell specificity.
ICE = Interleukin converting enzyme Caspase 1.
MICE = Min ICE.
PFP = Pore forming proteins.
PI = Propidium iodide, a DNA dye for excluding dead cells.
TRADD = TNF receptor associated death domain.
TRAIL = Tumor necrosis factor related apoptosis inducing ligand.
RAIL = Receptor of TRAIL.
TRAMP = TNF receptor apoptosis mediated protein.
Cell Proliferation and Apoptosis:
There is a relationship between tumor formation and apoptosis. P53 activates several numbers of genes involved in cell cycle regulation. ADV’s E1B, a 19KD protein blocks P53 ‘s transcriptional activation. Bcl2 is an Oncogene identified it activation due to activation of few genes identified in tumors. It actually blocks the pathway of apoptosis. So Apoptosis inhibits tumorigenesis and Bcl2 prevents apoptosis. P53 has pleotropic phenotypic effects; it triggers growth arrest and induces apoptosis. RB and P53 are activated in multiple routes. One locus that influences both RB and P53 is INK4A-ARF. Its transcript is alternately spiced as P16NK4A in RB and p19ARF in P53.

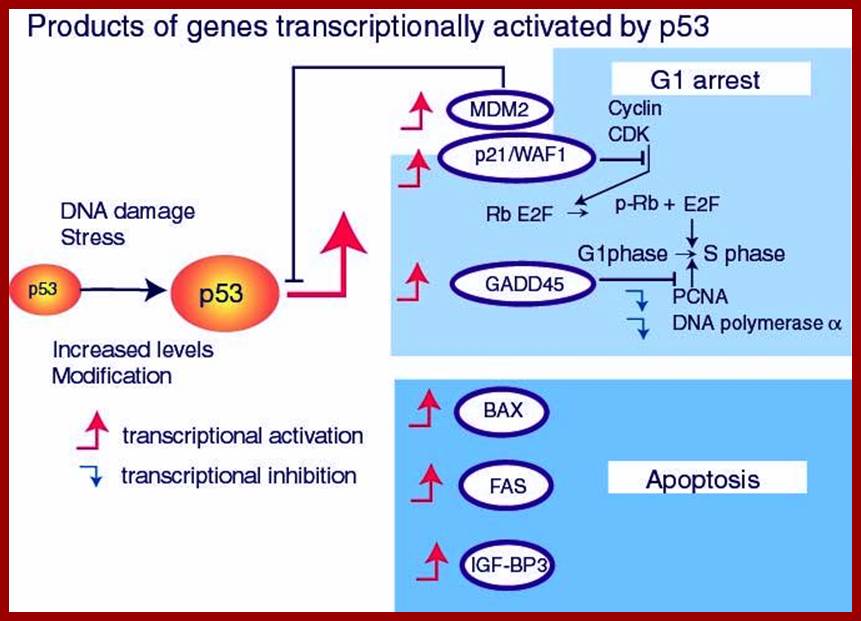
https://www.360zhyx.com
In human cancers deletion of INK4A-ARF is common which eliminates both P16INK4A and P19ARF, thus cell loses the ability to block tumor formation. P16INK4A inhibits cdk4/6 kinase, so it prevents the kinase from phosphorylation RB. In the absence of it cell cycle progression is halted in its track. The activity of INK4A is often disabled because of several point mutations in human tumors. P19ARF antagonizes Mdm2, which leads to stabilization of p53. Thus p19ARF acts as a tumor suppressor by inhibiting the inhibitor of P53. P19 may promote the degradation of Mdm2 or directly blocks interaction with p53. P19ARF arrests the cell cycle in p53 dependent manner. Loss of PARF and p53 has the same effect.
C-Myc and E1 (ADV) act via p19ARF to activate p53 dependent activation. P53 is often phosphorylated at serine residues (S6, S9, S37 etc). P53 is also acetylated at different positions. Ionizing radiations activate the kinase ATM, which phosphorylates S13 and S33 and S376 and L382. Such modifications may affect the stability, oligomerization, DNA binding, binding to other proteins. So p53 acts as a molecular sensor.